miRDB: an online database for prediction of functional microRNA targets
- PMID: 31504780
- PMCID: PMC6943051
- DOI: 10.1093/nar/gkz757
miRDB: an online database for prediction of functional microRNA targets
Abstract
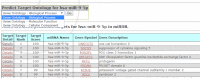
MicroRNAs (miRNAs) are small noncoding RNAs that act as master regulators in many biological processes. miRNAs function mainly by downregulating the expression of their gene targets. Thus, accurate prediction of miRNA targets is critical for characterization of miRNA functions. To this end, we have developed an online database, miRDB, for miRNA target prediction and functional annotations. Recently, we have performed major updates for miRDB. Specifically, by employing an improved algorithm for miRNA target prediction, we now present updated transcriptome-wide target prediction data in miRDB, including 3.5 million predicted targets regulated by 7000 miRNAs in five species. Further, we have implemented the new prediction algorithm into a web server, allowing custom target prediction with user-provided sequences. Another new database feature is the prediction of cell-specific miRNA targets. miRDB now hosts the expression profiles of over 1000 cell lines and presents target prediction data that are tailored for specific cell models. At last, a new web query interface has been added to miRDB for prediction of miRNA functions by integrative analysis of target prediction and Gene Ontology data. All data in miRDB are freely accessible at http://mirdb.org.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
miRDB: an online resource for microRNA target prediction and functional annotations.Nucleic Acids Res. 2015 Jan;43(Database issue):D146-52. doi: 10.1093/nar/gku1104. Epub 2014 Nov 5. Nucleic Acids Res. 2015. PMID: 25378301 Free PMC article.
-
Computational prediction of microRNA targets.Methods Mol Biol. 2010;667:283-95. doi: 10.1007/978-1-60761-811-9_19. Methods Mol Biol. 2010. PMID: 20827541
-
miRDB: a microRNA target prediction and functional annotation database with a wiki interface.RNA. 2008 Jun;14(6):1012-7. doi: 10.1261/rna.965408. Epub 2008 Apr 21. RNA. 2008. PMID: 18426918 Free PMC article.
-
Computational Prediction of MicroRNA Target Genes, Target Prediction Databases, and Web Resources.Methods Mol Biol. 2017;1617:109-122. doi: 10.1007/978-1-4939-7046-9_8. Methods Mol Biol. 2017. PMID: 28540680 Review.
-
Identifying miRNAs, targets and functions.Brief Bioinform. 2014 Jan;15(1):1-19. doi: 10.1093/bib/bbs075. Epub 2012 Nov 22. Brief Bioinform. 2014. PMID: 23175680 Free PMC article. Review.
Cited by
-
Identification of novel targets of miR-622 in hepatocellular carcinoma reveals common regulation of cooperating genes and outlines the oncogenic role of zinc finger CCHC-type containing 11.Neoplasia. 2021 May;23(5):502-514. doi: 10.1016/j.neo.2021.04.001. Epub 2021 Apr 24. Neoplasia. 2021. PMID: 33901943 Free PMC article.
-
Whole-Transcriptome RNA Sequencing Reveals Significant Differentially Expressed mRNAs, miRNAs, and lncRNAs and Related Regulating Biological Pathways in the Peripheral Blood of COVID-19 Patients.Mediators Inflamm. 2021 Apr 1;2021:6635925. doi: 10.1155/2021/6635925. eCollection 2021. Mediators Inflamm. 2021. PMID: 33833618 Free PMC article.
-
MicroRNA Expression Profile in TSC Cell Lines and the Impact of mTOR Inhibitor.Int J Mol Sci. 2022 Nov 21;23(22):14493. doi: 10.3390/ijms232214493. Int J Mol Sci. 2022. PMID: 36430972 Free PMC article.
-
miR-148a-3p and DDX6 functional link promotes survival of myeloid leukemia cells.Blood Adv. 2023 Aug 8;7(15):3846-3861. doi: 10.1182/bloodadvances.2022008123. Blood Adv. 2023. PMID: 36322827 Free PMC article.
-
isomiRTar: a comprehensive portal of pan-cancer 5'-isomiR targeting.PeerJ. 2022 Oct 17;10:e14205. doi: 10.7717/peerj.14205. eCollection 2022. PeerJ. 2022. PMID: 36275459 Free PMC article.
References
-
- Ambros V. The functions of animal microRNAs. Nature. 2004; 431:350–355. - PubMed
-
- Miska E.A. How microRNAs control cell division, differentiation and death. Curr. Opin. Genet. Dev. 2005; 15:563–568. - PubMed
-
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M.. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005; 433:769–773. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

