Novel ribozymes: discovery, catalytic mechanisms, and the quest to understand biological function
- PMID: 31504786
- PMCID: PMC6765202
- DOI: 10.1093/nar/gkz737
Novel ribozymes: discovery, catalytic mechanisms, and the quest to understand biological function
Abstract
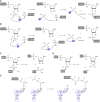
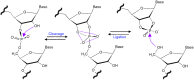
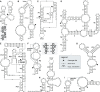
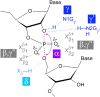
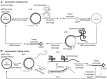
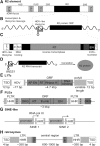
Small endonucleolytic ribozymes promote the self-cleavage of their own phosphodiester backbone at a specific linkage. The structures of and the reactions catalysed by members of individual families have been studied in great detail in the past decades. In recent years, bioinformatics studies have uncovered a considerable number of new examples of known catalytic RNA motifs. Importantly, entirely novel ribozyme classes were also discovered, for most of which both structural and biochemical information became rapidly available. However, for the majority of the new ribozymes, which are found in the genomes of a variety of species, a biological function remains elusive. Here, we concentrate on the different approaches to find catalytic RNA motifs in sequence databases. We summarize the emerging principles of RNA catalysis as observed for small endonucleolytic ribozymes. Finally, we address the biological functions of those ribozymes, where relevant information is available and common themes on their cellular activities are emerging. We conclude by speculating on the possibility that the identification and characterization of proteins that we hypothesize to be endogenously associated with catalytic RNA might help in answering the ever-present question of the biological function of the growing number of genomically encoded, small endonucleolytic ribozymes.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
The Varkud Satellite Ribozyme: A Thirty-Year Journey through Biochemistry, Crystallography, and Computation.Acc Chem Res. 2021 Jun 1;54(11):2591-2602. doi: 10.1021/acs.accounts.1c00052. Epub 2021 May 11. Acc Chem Res. 2021. PMID: 33974386 Review.
-
Inverse RNA Folding Workflow to Design and Test Ribozymes that Include Pseudoknots.Methods Mol Biol. 2021;2167:113-143. doi: 10.1007/978-1-0716-0716-9_8. Methods Mol Biol. 2021. PMID: 32712918
-
A combinatorial method to isolate short ribozymes from complex ribozyme libraries.Nucleic Acids Res. 2020 Nov 18;48(20):e116. doi: 10.1093/nar/gkaa834. Nucleic Acids Res. 2020. PMID: 33035338 Free PMC article.
-
Generation and development of RNA ligase ribozymes with modular architecture through "design and selection".Molecules. 2010 Aug 26;15(9):5850-65. doi: 10.3390/molecules15095850. Molecules. 2010. PMID: 22273983 Free PMC article. Review.
-
High-Throughput Analysis and Engineering of Ribozymes and Deoxyribozymes by Sequencing.Acc Chem Res. 2020 Dec 15;53(12):2903-2912. doi: 10.1021/acs.accounts.0c00546. Epub 2020 Nov 9. Acc Chem Res. 2020. PMID: 33164502
Cited by
-
Self-cleaving ribozymes: substrate specificity and synthetic biology applications.RSC Chem Biol. 2021 Jul 2;2(5):1370-1383. doi: 10.1039/d0cb00207k. eCollection 2021 Oct 7. RSC Chem Biol. 2021. PMID: 34704043 Free PMC article. Review.
-
Satellite-Like W-Elements: Repetitive, Transcribed, and Putative Mobile Genetic Factors with Potential Roles for Biology and Evolution of Schistosoma mansoni.Genome Biol Evol. 2021 Oct 1;13(10):evab204. doi: 10.1093/gbe/evab204. Genome Biol Evol. 2021. PMID: 34469545 Free PMC article.
-
Uncovered diversity of infectious circular RNAs: A new paradigm for the minimal parasites?Npj Viruses. 2024 Apr 18;2(1):13. doi: 10.1038/s44298-024-00023-7. Npj Viruses. 2024. PMID: 40295681 Free PMC article. Review.
-
Oligonucleotide-Based Approaches to Inhibit Dengue Virus Replication.Molecules. 2021 Feb 11;26(4):956. doi: 10.3390/molecules26040956. Molecules. 2021. PMID: 33670247 Free PMC article. Review.
-
The function of twister ribozyme variants in non-LTR retrotransposition in Schistosoma mansoni.Nucleic Acids Res. 2021 Oct 11;49(18):10573-10588. doi: 10.1093/nar/gkab818. Nucleic Acids Res. 2021. PMID: 34551436 Free PMC article.
References
-
- Cech T.R., Zaug A.J., Grabowski P.J.. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1981; 27:487–496. - PubMed
-
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S.. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983; 35:849–857. - PubMed
-
- Peebles C.L., Perlman P.S., Mecklenburg K.L., Petrillo M.L., Tabor J.H., Jarrell K.A., Cheng H.L.. A self-splicing RNA excises an intron lariat. Cell. 1986; 44:213–223. - PubMed
-
- van der Veen R., Arnberg A.C., van der Horst G., Bonen L., Tabak H.F., Grivell L.A.. Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell. 1986; 44:225–234. - PubMed
-
- Lilley D.M. Structure, folding and mechanisms of ribozymes. Curr. Opin. Struct. Biol. 2005; 15:313–323. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

