Alternative ORFs and small ORFs: shedding light on the dark proteome
- PMID: 31504789
- PMCID: PMC7026640
- DOI: 10.1093/nar/gkz734
Alternative ORFs and small ORFs: shedding light on the dark proteome
Abstract
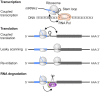
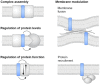
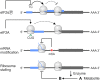
Traditional annotation of protein-encoding genes relied on assumptions, such as one open reading frame (ORF) encodes one protein and minimal lengths for translated proteins. With the serendipitous discoveries of translated ORFs encoded upstream and downstream of annotated ORFs, from alternative start sites nested within annotated ORFs and from RNAs previously considered noncoding, it is becoming clear that these initial assumptions are incorrect. The findings have led to the realization that genetic information is more densely coded and that the proteome is more complex than previously anticipated. As such, interest in the identification and characterization of the previously ignored 'dark proteome' is increasing, though we note that research in eukaryotes and bacteria has largely progressed in isolation. To bridge this gap and illustrate exciting findings emerging from studies of the dark proteome, we highlight recent advances in both eukaryotic and bacterial cells. We discuss progress in the detection of alternative ORFs as well as in the understanding of functions and the regulation of their expression and posit questions for future work.
Published by Oxford University Press on behalf of Nucleic Acids Research 2019.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

