mTOR-regulated U2af1 tandem exon splicing specifies transcriptome features for translational control
- PMID: 31504847
- PMCID: PMC6821156
- DOI: 10.1093/nar/gkz761
mTOR-regulated U2af1 tandem exon splicing specifies transcriptome features for translational control
Abstract
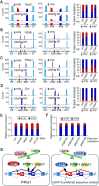
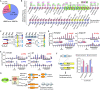
U2 auxiliary factor 1 (U2AF1) functions in 3'-splice site selection during pre-mRNA processing. Alternative usage of duplicated tandem exons in U2AF1 produces two isoforms, U2AF1a and U2AF1b, but their functional differences are unappreciated due to their homology. Through integrative approaches of genome editing, customized-transcriptome profiling and crosslinking-mediated interactome analyses, we discovered that the expression of U2AF1 isoforms is controlled by mTOR and they exhibit a distinctive molecular profile for the splice site and protein interactomes. Mechanistic dissection of mutually exclusive alternative splicing events revealed that U2AF1 isoforms' inherent differential preferences of nucleotide sequences and their stoichiometry determine the 3'-splice site. Importantly, U2AF1a-driven transcriptomes feature alternative splicing events in the 5'-untranslated region (5'-UTR) that are favorable for translation. These findings unveil distinct roles of duplicated tandem exon-derived U2AF1 isoforms in the regulation of the transcriptome and suggest U2AF1a-driven 5'-UTR alternative splicing as a molecular mechanism of mTOR-regulated translational control.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Alternative splicing of U2AF1 reveals a shared repression mechanism for duplicated exons.Nucleic Acids Res. 2017 Jan 9;45(1):417-434. doi: 10.1093/nar/gkw733. Epub 2016 Aug 26. Nucleic Acids Res. 2017. PMID: 27566151 Free PMC article.
-
Elucidation of the aberrant 3' splice site selection by cancer-associated mutations on the U2AF1.Nat Commun. 2020 Sep 21;11(1):4744. doi: 10.1038/s41467-020-18559-6. Nat Commun. 2020. PMID: 32958768 Free PMC article.
-
Diversity of vertebrate splicing factor U2AF35: identification of alternatively spliced U2AF1 mRNAS.J Biol Chem. 2004 Jun 25;279(26):27039-49. doi: 10.1074/jbc.M402136200. Epub 2004 Apr 19. J Biol Chem. 2004. PMID: 15096518
-
U2AF1 in various neoplastic diseases and relevant targeted therapies for malignant cancers with complex mutations (Review).Oncol Rep. 2024 Jan;51(1):5. doi: 10.3892/or.2023.8664. Epub 2023 Nov 17. Oncol Rep. 2024. PMID: 37975232 Free PMC article. Review.
-
Alternative splicing of mutually exclusive exons--a review.Biosystems. 2013 Oct;114(1):31-8. doi: 10.1016/j.biosystems.2013.07.003. Epub 2013 Jul 11. Biosystems. 2013. PMID: 23850531 Review.
Cited by
-
The expression and significance of mTORC1 in diabetic retinopathy.BMC Ophthalmol. 2020 Jul 20;20(1):297. doi: 10.1186/s12886-020-01553-3. BMC Ophthalmol. 2020. PMID: 32689970 Free PMC article.
-
A systems-based approach to uterine fibroids identifies differential splicing associated with abnormal uterine bleeding.Commun Med (Lond). 2025 Jul 31;5(1):318. doi: 10.1038/s43856-025-01051-x. Commun Med (Lond). 2025. PMID: 40739398 Free PMC article.
-
Light regulates alternative splicing outcomes via the TOR kinase pathway.Cell Rep. 2021 Sep 7;36(10):109676. doi: 10.1016/j.celrep.2021.109676. Cell Rep. 2021. PMID: 34496244 Free PMC article.
-
mTOR-coordinated Post-Transcriptional Gene Regulations: from Fundamental to Pathogenic Insights.J Lipid Atheroscler. 2020 Jan;9(1):8-22. doi: 10.12997/jla.2020.9.1.8. Epub 2019 Oct 18. J Lipid Atheroscler. 2020. PMID: 32821719 Free PMC article. Review.
-
Exploring transcriptome plasticity at the intersection of cell signaling, metabolism and computational biology.Exp Mol Med. 2025 Aug 14. doi: 10.1038/s12276-025-01513-1. Online ahead of print. Exp Mol Med. 2025. PMID: 40804478 No abstract available.
References
-
- Shi Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 2017; 18:655–670. - PubMed
-
- Zamore P.D., Patton J.G., Green M.R.. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992; 355:609–614. - PubMed
-
- Singh R., Valcárcel J., Green M.R.. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995; 268:1173–1176. - PubMed
-
- Valcárcel J., Gaur R.K., Singh R., Green M.R.. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA [corrected]. Science. 1996; 273:1706–1709. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

