Identification of microRNAs involved in pathways which characterize the expression subtypes of NSCLC
- PMID: 31505091
- PMCID: PMC6887593
- DOI: 10.1002/1878-0261.12571
Identification of microRNAs involved in pathways which characterize the expression subtypes of NSCLC
Abstract
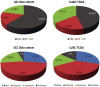
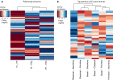
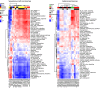
Dysregulation of microRNAs is a common mechanism in the development of lung cancer, but the relationship between microRNAs and expression subtypes in non-small-cell lung cancer (NSCLC) is poorly explored. Here, we analyzed microRNA expression from 241 NSCLC samples and correlated this with the expression subtypes of adenocarcinomas (AD) and squamous cell carcinomas (SCC) to identify microRNAs specific for each subtype. Gene set variation analysis and the hallmark gene set were utilized to calculate gene set scores specific for each sample, and these were further correlated with the expression of the subtype-specific microRNAs. In ADs, we identified nine aberrantly regulated microRNAs in the terminal respiratory unit (TRU), three in the proximal inflammatory (PI), and nine in the proximal proliferative subtype (PP). In SCCs, 1, 5, 5, and 9 microRNAs were significantly dysregulated in the basal, primitive, classical, and secretory subtypes, respectively. The subtype-specific microRNAs were highly correlated to specific gene sets, and a distinct pattern of biological processes with high immune activity for the AD PI and SCC secretory subtypes, and upregulation of cell cycle-related processes in AD PP, SCC primitive, and SCC classical subtypes were found. Several in silico predicted targets within the gene sets were identified for the subtype-specific microRNAs, underpinning the findings. The results were significantly validated in the LUAD (n = 492) and LUSC (n = 380) TCGA dataset (False discovery rates-corrected P-value < 0.05). Our study provides novel insight into how expression subtypes determined with discrete biological processes may be regulated by subtype-specific microRNAs. These results may have importance for the development of combinatory therapeutic strategies for lung cancer patients.
Keywords: adenocarcinoma; expression subtypes; microRNA; non-small-cell lung cancer; pathway; predicted target; squamous cell carcinoma.
© 2019 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Lung Adenocarcinoma and Squamous Cell Carcinoma Gene Expression Subtypes Demonstrate Significant Differences in Tumor Immune Landscape.J Thorac Oncol. 2017 Jun;12(6):943-953. doi: 10.1016/j.jtho.2017.03.010. Epub 2017 Mar 21. J Thorac Oncol. 2017. PMID: 28341226 Free PMC article.
-
Molecular subtypes of lung adenocarcinoma present distinct immune tumor microenvironments.Cancer Sci. 2024 Jun;115(6):1763-1777. doi: 10.1111/cas.16154. Epub 2024 Mar 25. Cancer Sci. 2024. PMID: 38527308 Free PMC article.
-
MicroRNA dysregulational synergistic network: discovering microRNA dysregulatory modules across subtypes in non-small cell lung cancers.BMC Bioinformatics. 2018 Dec 21;19(Suppl 20):504. doi: 10.1186/s12859-018-2536-0. BMC Bioinformatics. 2018. PMID: 30577741 Free PMC article.
-
Downregulation of HOXA3 in lung adenocarcinoma and its relevant molecular mechanism analysed by RT-qPCR, TCGA and in silico analysis.Int J Oncol. 2018 Oct;53(4):1557-1579. doi: 10.3892/ijo.2018.4508. Epub 2018 Jul 30. Int J Oncol. 2018. PMID: 30066858 Free PMC article.
-
Expression of miRNAs in non-small-cell lung carcinomas and their association with clinicopathological features.Tumour Biol. 2015 Mar;36(3):1603-12. doi: 10.1007/s13277-014-2755-6. Epub 2014 Nov 11. Tumour Biol. 2015. PMID: 25384507
Cited by
-
Identification of biomarkers associated with diagnosis of postmenopausal osteoporosis patients based on bioinformatics and machine learning.Front Genet. 2023 Jul 3;14:1198417. doi: 10.3389/fgene.2023.1198417. eCollection 2023. Front Genet. 2023. PMID: 37465165 Free PMC article.
-
MiRNA expression profiling in adenocarcinoma and squamous cell lung carcinoma reveals both common and specific deregulated microRNAs.Medicine (Baltimore). 2022 Aug 19;101(33):e30027. doi: 10.1097/MD.0000000000030027. Medicine (Baltimore). 2022. PMID: 35984198 Free PMC article.
-
MicroRNA-326 impairs chemotherapy resistance in non small cell lung cancer by suppressing histone deacetylase SIRT1-mediated HIF1α and elevating VEGFA.Bioengineered. 2022 Mar;13(3):5685-5699. doi: 10.1080/21655979.2021.1993718. Bioengineered. 2022. PMID: 34696659 Free PMC article.
-
HPV-related methylation-based reclassification and risk stratification of cervical cancer.Mol Oncol. 2020 Sep;14(9):2124-2141. doi: 10.1002/1878-0261.12709. Epub 2020 Jun 2. Mol Oncol. 2020. PMID: 32408396 Free PMC article.
-
Exploring the Role of Circulating Cell-Free RNA in the Development of Colorectal Cancer.Int J Mol Sci. 2023 Jul 3;24(13):11026. doi: 10.3390/ijms241311026. Int J Mol Sci. 2023. PMID: 37446204 Free PMC article. Review.
References
-
- Calin GA and Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6, 857–866. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

