Tales from topographic oceans: topologically associated domains and cancer
- PMID: 31505466
- PMCID: PMC7664306
- DOI: 10.1530/ERC-19-0348
Tales from topographic oceans: topologically associated domains and cancer
Abstract
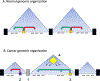
The 3D organization of the genome within the cell nucleus has come into sharp focus over the last decade. This has largely arisen because of the application of genomic approaches that have revealed numerous levels of genomic and chromatin interactions, including topologically associated domains (TADs). The current review examines how these domains were identified, are organized, how their boundaries arise and are regulated, and how genes within TADs are coordinately regulated. There are many examples of the disruption to TAD structure in cancer and the altered regulation, structure and function of TADs are discussed in the context of hormone responsive cancers, including breast, prostate and ovarian cancer. Finally, some aspects of the statistical insight and computational skills required to interrogate TAD organization are considered and future directions discussed.
Keywords: CTCF; breast cancer; cohesin; enhancer; ovarian cancer; prostate cancer; topologically associated domain.
Figures
Similar articles
-
TAD-dependent sub-TAD is required for enhancer-promoter interaction enabling the β-globin transcription.FASEB J. 2024 Nov 30;38(22):e70181. doi: 10.1096/fj.202401526RR. FASEB J. 2024. PMID: 39545685 Free PMC article.
-
Clustered CTCF binding is an evolutionary mechanism to maintain topologically associating domains.Genome Biol. 2020 Jan 7;21(1):5. doi: 10.1186/s13059-019-1894-x. Genome Biol. 2020. PMID: 31910870 Free PMC article.
-
CTCF modulates allele-specific sub-TAD organization and imprinted gene activity at the mouse Dlk1-Dio3 and Igf2-H19 domains.Genome Biol. 2019 Dec 12;20(1):272. doi: 10.1186/s13059-019-1896-8. Genome Biol. 2019. PMID: 31831055 Free PMC article.
-
A tour of 3D genome with a focus on CTCF.Semin Cell Dev Biol. 2019 Jun;90:4-11. doi: 10.1016/j.semcdb.2018.07.020. Epub 2018 Jul 23. Semin Cell Dev Biol. 2019. PMID: 30031214 Review.
-
Chromatin Architecture in the Fly: Living without CTCF/Cohesin Loop Extrusion?: Alternating Chromatin States Provide a Basis for Domain Architecture in Drosophila.Bioessays. 2019 Sep;41(9):e1900048. doi: 10.1002/bies.201900048. Epub 2019 Jul 1. Bioessays. 2019. PMID: 31264253 Review.
Cited by
-
Reduced NCOR2 expression accelerates androgen deprivation therapy failure in prostate cancer.Cell Rep. 2021 Dec 14;37(11):110109. doi: 10.1016/j.celrep.2021.110109. Cell Rep. 2021. PMID: 34910907 Free PMC article.
-
A functional mechanism for a non-coding variant near AGTR2 associated with risk for preterm birth.BMC Med. 2023 Jul 17;21(1):258. doi: 10.1186/s12916-023-02973-w. BMC Med. 2023. PMID: 37455310 Free PMC article.
References
-
- ARCECI RJ & GROSS PR 1980. Histone gene expression: progeny of isolated early blastomeres in culture make the same change as in the embryo. Science, 209, 607–9. - PubMed
-
- BELL AC, WEST AG & FELSENFELD G 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell, 98, 387–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

