Antiviral Effect of Lithium Chloride and Diammonium Glycyrrhizinate on Porcine Deltacoronavirus In Vitro
- PMID: 31505777
- PMCID: PMC6789623
- DOI: 10.3390/pathogens8030144
Antiviral Effect of Lithium Chloride and Diammonium Glycyrrhizinate on Porcine Deltacoronavirus In Vitro
Abstract
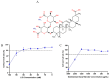
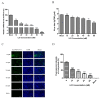
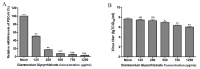
Porcine deltacoronavirus (PDCoV) is an emerging global swine virus that has a propensity for interspecies transmission. It was identified in Hong Kong in 2012. Given that neither specific antiviral drugs nor vaccines are available for newly emerging porcine deltacoronavirus, searching for effective antiviral drugs is a high priority. In this study, lithium chloride (LiCl) and diammonium glycyrrhizinate (DG), which are host-acting antivirals (HAAs), were tested against PDCoV. We found that LiCl and DG inhibited PDCoV replication in LLC-PK1 cells in a dose-dependent manner. The antiviral effects of LiCl and DG occurred at the early stage of PDCoV replication, and DG also inhibited virus attachment to the cells. Moreover, both drugs inhibited PDCoV-induced apoptosis in LLC-PK1 cells. This study suggests LiCl and DG as new drugs for the treatment of PDCoV infection.
Keywords: PDCoV; apoptosis; diammonium glycyrrhizinate; lithium chloride.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Ergosterol peroxide exhibits antiviral and immunomodulatory abilities against porcine deltacoronavirus (PDCoV) via suppression of NF-κB and p38/MAPK signaling pathways in vitro.Int Immunopharmacol. 2021 Apr;93:107317. doi: 10.1016/j.intimp.2020.107317. Epub 2021 Jan 22. Int Immunopharmacol. 2021. PMID: 33493866 Free PMC article.
-
Bile acids LCA and CDCA inhibited porcine deltacoronavirus replication in vitro.Vet Microbiol. 2021 Jun;257:109097. doi: 10.1016/j.vetmic.2021.109097. Epub 2021 Apr 28. Vet Microbiol. 2021. PMID: 33933854
-
Chlorogenic acid inhibits porcine deltacoronavirus release by targeting apoptosis.Int Immunopharmacol. 2024 Jan 25;127:111359. doi: 10.1016/j.intimp.2023.111359. Epub 2023 Dec 14. Int Immunopharmacol. 2024. PMID: 38101217
-
An Updated Review of Porcine Deltacoronavirus in Terms of Prevalence, Pathogenicity, Pathogenesis and Antiviral Strategy.Front Vet Sci. 2022 Jan 13;8:811187. doi: 10.3389/fvets.2021.811187. eCollection 2021. Front Vet Sci. 2022. PMID: 35097055 Free PMC article. Review.
-
Porcine deltacoronavirus: Overview of infection dynamics, diagnostic methods, prevalence and genetic evolution.Virus Res. 2016 Dec 2;226:71-84. doi: 10.1016/j.virusres.2016.05.028. Epub 2016 Jun 4. Virus Res. 2016. PMID: 27270129 Free PMC article. Review.
Cited by
-
Antiviral effects of ergosterol peroxide in a pig model of porcine deltacoronavirus (PDCoV) infection involves modulation of apoptosis and tight junction in the small intestine.Vet Res. 2021 Jun 14;52(1):86. doi: 10.1186/s13567-021-00955-5. Vet Res. 2021. PMID: 34127062 Free PMC article.
-
Does Lithium Deserve a Place in the Treatment Against COVID-19? A Preliminary Observational Study in Six Patients, Case Report.Front Pharmacol. 2020 Aug 27;11:557629. doi: 10.3389/fphar.2020.557629. eCollection 2020. Front Pharmacol. 2020. PMID: 32973537 Free PMC article.
-
A Review of the Antiviral Activities of Glycyrrhizic Acid, Glycyrrhetinic Acid and Glycyrrhetinic Acid Monoglucuronide.Pharmaceuticals (Basel). 2023 Apr 23;16(5):641. doi: 10.3390/ph16050641. Pharmaceuticals (Basel). 2023. PMID: 37242424 Free PMC article. Review.
-
Inhibition of porcine deltacoronavirus entry and replication by Cepharanthine.Virus Res. 2024 Feb;340:199303. doi: 10.1016/j.virusres.2023.199303. Epub 2023 Dec 30. Virus Res. 2024. PMID: 38145807 Free PMC article.
-
Porcine Deltacoronavirus (PDCoV) Entry into PK-15 Cells by Caveolae-Mediated Endocytosis.Viruses. 2022 Feb 28;14(3):496. doi: 10.3390/v14030496. Viruses. 2022. PMID: 35336903 Free PMC article.
References
-
- Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., Lau J.H.N., Bai R., Teng J.L.L., Tsang C.C.C., Wang M. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavir. J. Virol. 2012;86:3995. doi: 10.1128/JVI.06540-11. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

