SWEET Gene Family in Medicago truncatula: Genome-Wide Identification, Expression and Substrate Specificity Analysis
- PMID: 31505820
- PMCID: PMC6783836
- DOI: 10.3390/plants8090338
SWEET Gene Family in Medicago truncatula: Genome-Wide Identification, Expression and Substrate Specificity Analysis
Abstract
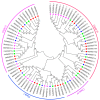
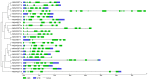
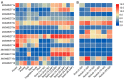
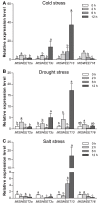
SWEET (Sugars Will Eventually be Exported Transporter) proteins mediate the translocation of sugars across cell membranes and play crucial roles in plant growth and development as well as stress responses. In this study, a total of 25 SWEET genes were identified from the Medicago truncatula genome and were divided into four clades based on the phylogenetic analysis. The MtSWEET genes are distributed unevenly on the M. truncatula chromosomes, and eight and 12 MtSWEET genes are segmentally and tandemly duplicated, respectively. Most MtSWEET genes contain five introns and encode proteins with seven transmembrane helices (TMHs). Besides, nearly all MtSWEET proteins have relatively conserved membrane domains, and contain conserved active sites. Analysis of microarray data showed that some MtSWEET genes are specifically expressed in disparate developmental stages or tissues, such as flowers, developing seeds and nodules. RNA-seq and qRT-PCR expression analysis indicated that many MtSWEET genes are responsive to various abiotic stresses such as cold, drought, and salt treatments. Functional analysis of six selected MtSWEETs in yeast revealed that they possess diverse transport activities for sucrose, fructose, glucose, galactose, and mannose. These results provide new insights into the characteristics of the MtSWEET genes, which lay a solid foundation for further investigating their functional roles in the developmental processes and stress responses of M. truncatula.
Keywords: Medicago truncatula; SWEET; abiotic stress; evolution; expression analysis; sugar transport.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Genome-Wide Identification and Expression Analysis of the SWEET Gene Family in Annual Alfalfa (Medicago polymorpha).Plants (Basel). 2023 May 10;12(10):1948. doi: 10.3390/plants12101948. Plants (Basel). 2023. PMID: 37653865 Free PMC article.
-
In silico identification and expression analysis of superoxide dismutase (SOD) gene family in Medicago truncatula.3 Biotech. 2018 Aug;8(8):348. doi: 10.1007/s13205-018-1373-1. Epub 2018 Jul 30. 3 Biotech. 2018. PMID: 30073133 Free PMC article.
-
Genome-wide identification and characterization of R2R3-MYB genes in Medicago truncatula.Genet Mol Biol. 2019 Jul-Sep;42(3):611-623. doi: 10.1590/1678-4685-GMB-2018-0235. Epub 2019 Nov 14. Genet Mol Biol. 2019. PMID: 31188936 Free PMC article.
-
Genome-wide identification, characterization and expression analysis of HAK genes and decoding their role in responding to potassium deficiency and abiotic stress in Medicago truncatula.PeerJ. 2022 Sep 22;10:e14034. doi: 10.7717/peerj.14034. eCollection 2022. PeerJ. 2022. PMID: 36168431 Free PMC article.
-
Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance.IUBMB Life. 2015 Jul;67(7):461-71. doi: 10.1002/iub.1394. Epub 2015 Jul 15. IUBMB Life. 2015. PMID: 26179993 Review.
Cited by
-
Precise CRISPR-Cas9 Mediated Genome Editing in Super Basmati Rice for Resistance Against Bacterial Blight by Targeting the Major Susceptibility Gene.Front Plant Sci. 2020 Jun 12;11:575. doi: 10.3389/fpls.2020.00575. eCollection 2020. Front Plant Sci. 2020. PMID: 32595655 Free PMC article.
-
Genome-wide analysis of the SWEET gene family in Hemerocallis citrina and functional characterization of HcSWEET4a in response to salt stress.BMC Plant Biol. 2024 Jul 11;24(1):661. doi: 10.1186/s12870-024-05376-y. BMC Plant Biol. 2024. PMID: 38987684 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of SWEET Family Genes in Sweet Potato and Its Two Diploid Relatives.Int J Mol Sci. 2022 Dec 13;23(24):15848. doi: 10.3390/ijms232415848. Int J Mol Sci. 2022. PMID: 36555491 Free PMC article.
-
Role of Brassica rapa SWEET genes in the defense response to Plasmodiophora brassicae.Genes Genomics. 2024 Feb;46(2):253-261. doi: 10.1007/s13258-023-01486-3. Epub 2024 Jan 18. Genes Genomics. 2024. PMID: 38236352
-
Emerging Roles of SWEET Sugar Transporters in Plant Development and Abiotic Stress Responses.Cells. 2022 Apr 12;11(8):1303. doi: 10.3390/cells11081303. Cells. 2022. PMID: 35455982 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

