OsWRKY5 Promotes Rice Leaf Senescence via Senescence-Associated NAC and Abscisic Acid Biosynthesis Pathway
- PMID: 31505875
- PMCID: PMC6770167
- DOI: 10.3390/ijms20184437
OsWRKY5 Promotes Rice Leaf Senescence via Senescence-Associated NAC and Abscisic Acid Biosynthesis Pathway
Abstract
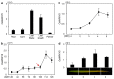
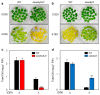
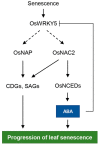
he onset of leaf senescence is triggered by external cues and internal factors such as phytohormones and signaling pathways involving transcription factors (TFs). Abscisic acid (ABA) strongly induces senescence and endogenous ABA levels are finely tuned by many senescence-associated TFs. Here, we report on the regulatory function of the senescence-induced TF OsWRKY5 TF in rice (Oryza sativa). OsWRKY5 expression was rapidly upregulated in senescing leaves, especially in yellowing sectors initiated by aging or dark treatment. A T-DNA insertion activation-tagged OsWRKY5-overexpressing mutant (termed oswrky5-D) promoted leaf senescence under natural and dark-induced senescence (DIS) conditions. By contrast, a T-DNA insertion oswrky5-knockdown mutant (termed oswrky5) retained leaf greenness during DIS. Reverse-transcription quantitative PCR (RT-qPCR) showed that OsWRKY5 upregulates the expression of genes controlling chlorophyll degradation and leaf senescence. Furthermore, RT-qPCR and yeast one-hybrid analysis demonstrated that OsWRKY5 indirectly upregulates the expression of senescence-associated NAM/ATAF1/2/CUC2 (NAC) genes including OsNAP and OsNAC2. Precocious leaf yellowing in the oswrky5-D mutant might be caused by elevated endogenous ABA concentrations resulting from upregulated expression of ABA biosynthesis genes OsNCED3, OsNCED4, and OsNCED5, indicating that OsWRKY is a positive regulator of ABA biosynthesis during leaf senescence. Furthermore, OsWRKY5 expression was suppressed by ABA treatment. Taken together, OsWRKY5 is a positive regulator of leaf senescence that upregulates senescence-induced NAC, ABA biosynthesis, and chlorophyll degradation genes.
Keywords: NAC; OsWRKY; abscisic acid (ABA); leaf senescence; rice.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
A Rice NAC Transcription Factor Promotes Leaf Senescence via ABA Biosynthesis.Plant Physiol. 2017 Jul;174(3):1747-1763. doi: 10.1104/pp.17.00542. Epub 2017 May 12. Plant Physiol. 2017. PMID: 28500268 Free PMC article.
-
Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling.J Exp Bot. 2019 May 9;70(10):2699-2715. doi: 10.1093/jxb/erz095. J Exp Bot. 2019. PMID: 30825376 Free PMC article.
-
OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice.Proc Natl Acad Sci U S A. 2014 Jul 8;111(27):10013-8. doi: 10.1073/pnas.1321568111. Epub 2014 Jun 20. Proc Natl Acad Sci U S A. 2014. PMID: 24951508 Free PMC article.
-
Abiotic Stresses Intervene with ABA Signaling to Induce Destructive Metabolic Pathways Leading to Death: Premature Leaf Senescence in Plants.Int J Mol Sci. 2019 Jan 10;20(2):256. doi: 10.3390/ijms20020256. Int J Mol Sci. 2019. PMID: 30634648 Free PMC article. Review.
-
Current Understanding of Leaf Senescence in Rice.Int J Mol Sci. 2021 Apr 26;22(9):4515. doi: 10.3390/ijms22094515. Int J Mol Sci. 2021. PMID: 33925978 Free PMC article. Review.
Cited by
-
Light Deficiency Inhibits Growth by Affecting Photosynthesis Efficiency as well as JA and Ethylene Signaling in Endangered Plant Magnolia sinostellata.Plants (Basel). 2021 Oct 22;10(11):2261. doi: 10.3390/plants10112261. Plants (Basel). 2021. PMID: 34834626 Free PMC article.
-
Rice Basic Helix-Loop-Helix 079 (OsbHLH079) Delays Leaf Senescence by Attenuating ABA Signaling.Rice (N Y). 2023 Dec 13;16(1):60. doi: 10.1186/s12284-023-00673-w. Rice (N Y). 2023. PMID: 38093151 Free PMC article.
-
Molecular and Physiological Perspectives of Abscisic Acid Mediated Drought Adjustment Strategies.Plants (Basel). 2021 Dec 15;10(12):2769. doi: 10.3390/plants10122769. Plants (Basel). 2021. PMID: 34961239 Free PMC article. Review.
-
ZIP Genes Are Involved in the Retransfer of Zinc Ions during the Senescence of Zinc-Deficient Rice Leaves.Int J Mol Sci. 2023 Sep 12;24(18):13989. doi: 10.3390/ijms241813989. Int J Mol Sci. 2023. PMID: 37762290 Free PMC article.
-
Know when and how to die: gaining insights into the molecular regulation of leaf senescence.Physiol Mol Biol Plants. 2022 Aug;28(8):1515-1534. doi: 10.1007/s12298-022-01224-1. Epub 2022 Aug 30. Physiol Mol Biol Plants. 2022. PMID: 36389097 Free PMC article. Review.
References
-
- Guo Y., Gan S. Leaf senescence: Signals, execution, and regulation. Curr. Top. Dev. Biol. 2005;71:83–112. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

