Biophysical Insight on the Membrane Insertion of an Arginine-Rich Cell-Penetrating Peptide
- PMID: 31505894
- PMCID: PMC6769507
- DOI: 10.3390/ijms20184441
Biophysical Insight on the Membrane Insertion of an Arginine-Rich Cell-Penetrating Peptide
Abstract
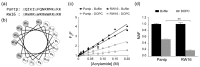
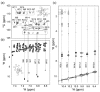
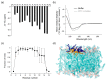
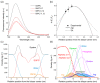
Cell-penetrating peptides (CPPs) are short peptides that can translocate and transport cargoes into the intracellular milieu by crossing biological membranes. The mode of interaction and internalization of cell-penetrating peptides has long been controversial. While their interaction with anionic membranes is quite well understood, the insertion and behavior of CPPs in zwitterionic membranes, a major lipid component of eukaryotic cell membranes, is poorly studied. Herein, we investigated the membrane insertion of RW16 into zwitterionic membranes, a versatile CPP that also presents antibacterial and antitumor activities. Using complementary approaches, including NMR spectroscopy, fluorescence spectroscopy, circular dichroism, and molecular dynamic simulations, we determined the high-resolution structure of RW16 and measured its membrane insertion and orientation properties into zwitterionic membranes. Altogether, these results contribute to explaining the versatile properties of this peptide toward zwitterionic lipids.
Keywords: NMR; cell-penetrating peptide; lipid model systems; membrane biophysics; molecular dynamics; peptide–lipid interaction.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Langel Ü. Methods for Structural Studies of CPPs. CPP, Cell-Penetrating Peptides. Springer Singapore; Singapore: 2019. pp. 289–323.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

