Germline Maintenance Through the Multifaceted Activities of GLH/Vasa in Caenorhabditis elegans P Granules
- PMID: 31506335
- PMCID: PMC6827368
- DOI: 10.1534/genetics.119.302670
Germline Maintenance Through the Multifaceted Activities of GLH/Vasa in Caenorhabditis elegans P Granules
Abstract
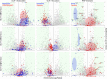
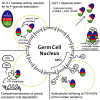
Vasa homologs are ATP-dependent DEAD-box helicases, multipotency factors, and critical components that specify and protect the germline. They regulate translation, amplify piwi-interacting RNAs (piRNAs), and act as RNA solvents; however, the limited availability of mutagenesis-derived alleles and their wide range of phenotypes have complicated their analysis. Now, with clustered regularly interspaced short palindromic repeats (CRISPR/Cas9), these limitations can be mitigated to determine why protein domains have been lost or retained throughout evolution. Here, we define the functional motifs of GLH-1/Vasa in Caenorhabditis elegans using 28 endogenous, mutant alleles. We show that GLH-1's helicase activity is required to retain its association with P granules. GLH-1 remains in P granules when changes are made outside of the helicase and flanking domains, but fertility is still compromised. Removal of the glycine-rich repeats from GLH proteins progressively diminishes P-granule wetting-like interactions at the nuclear periphery. Mass spectrometry of GLH-1-associated proteins implies conservation of a transient piRNA-amplifying complex, and reveals a novel affinity between GLH-1 and three structurally conserved PCI (26S Proteasome Lid, COP9, and eIF3) complexes or "zomes," along with a reciprocal aversion for assembled ribosomes and the 26S proteasome. These results suggest that P granules compartmentalize the cytoplasm to exclude large protein assemblies, effectively shielding associated transcripts from translation and associated proteins from turnover. Within germ granules, Vasa homologs may act as solvents, ensuring mRNA accessibility by small RNA surveillance and amplification pathways, and facilitating mRNA export through germ granules to initiate translation.
Keywords: C. elegans; DDX4; GLH-1; P granules; PCI complex; Vasa; germ granules; germline.
Copyright © 2019 by the Genetics Society of America.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

