Global invasion history of the agricultural pest butterfly Pieris rapae revealed with genomics and citizen science
- PMID: 31506352
- PMCID: PMC6778179
- DOI: 10.1073/pnas.1907492116
Global invasion history of the agricultural pest butterfly Pieris rapae revealed with genomics and citizen science
Abstract
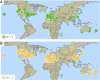
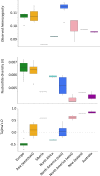
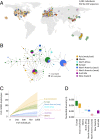
The small cabbage white butterfly, Pieris rapae, is a major agricultural pest of cruciferous crops and has been introduced to every continent except South America and Antarctica as a result of human activities. In an effort to reconstruct the near-global invasion history of P. rapae, we developed a citizen science project, the "Pieris Project," and successfully amassed thousands of specimens from 32 countries worldwide. We then generated and analyzed nuclear (double-digest restriction site-associated DNA fragment procedure [ddRAD]) and mitochondrial DNA sequence data for these samples to reconstruct and compare different global invasion history scenarios. Our results bolster historical accounts of the global spread and timing of P. rapae introductions. We provide molecular evidence supporting the hypothesis that the ongoing divergence of the European and Asian subspecies of P. rapae (∼1,200 y B.P.) coincides with the diversification of brassicaceous crops and the development of human trade routes such as the Silk Route (Silk Road). The further spread of P. rapae over the last ∼160 y was facilitated by human movement and trade, resulting in an almost linear series of at least 4 founding events, with each introduced population going through a severe bottleneck and serving as the source for the next introduction. Management efforts of this agricultural pest may need to consider the current existence of multiple genetically distinct populations. Finally, the international success of the Pieris Project demonstrates the power of the public to aid scientists in collections-based research addressing important questions in invasion biology, and in ecology and evolutionary biology more broadly.
Keywords: agricultural pest; approximate Bayesian computation; citizen science; genomics; invasive.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Mitochondrial DNA sequence variation of the swallowtail butterfly, Papilio xuthus, and the cabbage butterfly, Pieris rapae.Biochem Genet. 2009 Apr;47(3-4):165-78. doi: 10.1007/s10528-008-9214-2. Epub 2009 Jan 29. Biochem Genet. 2009. PMID: 19184408
-
Genomic adaptation to agricultural environments: cabbage white butterflies (Pieris rapae) as a case study.BMC Genomics. 2017 May 26;18(1):412. doi: 10.1186/s12864-017-3787-2. BMC Genomics. 2017. PMID: 28549454 Free PMC article.
-
Population genomics provides insights into the genetic diversity and adaptation of the Pieris rapae in China.PLoS One. 2023 Nov 16;18(11):e0294521. doi: 10.1371/journal.pone.0294521. eCollection 2023. PLoS One. 2023. PMID: 37972203 Free PMC article.
-
Biological invasions in agricultural settings: insights from evolutionary biology and population genetics.C R Biol. 2011 Mar;334(3):237-46. doi: 10.1016/j.crvi.2010.12.008. Epub 2011 Feb 1. C R Biol. 2011. PMID: 21377619 Review.
-
Social Inclusion and Pest Management: a Route for Improved Food Production and well-being in the Global South.Neotrop Entomol. 2024 Oct;53(5):1009-1012. doi: 10.1007/s13744-024-01179-w. Epub 2024 Jul 16. Neotrop Entomol. 2024. PMID: 39012615 Review.
Cited by
-
Migratory behaviour is positively associated with genetic diversity in butterflies.Mol Ecol. 2023 Feb;32(3):560-574. doi: 10.1111/mec.16770. Epub 2022 Nov 23. Mol Ecol. 2023. PMID: 36336800 Free PMC article.
-
Characterization of a cell death-inducing endonuclease-like venom protein from the parasitoid wasp Pteromalus puparum (Hymenoptera: Pteromalidae).Pest Manag Sci. 2021 Jan;77(1):224-233. doi: 10.1002/ps.6011. Epub 2020 Aug 6. Pest Manag Sci. 2021. PMID: 32673424 Free PMC article.
-
Expanding insect pollinators in the Anthropocene.Biol Rev Camb Philos Soc. 2021 Dec;96(6):2755-2770. doi: 10.1111/brv.12777. Epub 2021 Jul 21. Biol Rev Camb Philos Soc. 2021. PMID: 34288353 Free PMC article. Review.
-
Darwinian genomics and diversity in the tree of life.Proc Natl Acad Sci U S A. 2022 Jan 25;119(4):e2115644119. doi: 10.1073/pnas.2115644119. Proc Natl Acad Sci U S A. 2022. PMID: 35042807 Free PMC article.
-
Sympatric Pieris butterfly species exhibit a high conservation of chemoreceptors.Front Cell Neurosci. 2023 May 11;17:1155405. doi: 10.3389/fncel.2023.1155405. eCollection 2023. Front Cell Neurosci. 2023. PMID: 37252192 Free PMC article.
References
-
- Meyerson L. A., Mooney H. A., Invasive alien species in an era of globalization. Front. Ecol. Environ. 5, 199–208 (2007).
-
- Westphal M. I., Browne M., MacKinnon K., Noble I., The link between international trade and the global distribution of invasive alien species. Biol. Invasions 10, 391–398 (2008).
-
- Chen Y. H., Crop domestication, global human-mediated migration, and the unresolved role of geography in pest control. Elem. Sci. Anth. 4, 000106 (2016).
-
- Estoup A., Guillemaud T., Reconstructing routes of invasion using genetic data: Why, how and so what? Mol. Ecol. 19, 4113–4130 (2010). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

