Large-scale integrated analysis of ovarian cancer tumors and cell lines identifies an individualized gene expression signature for predicting response to platinum-based chemotherapy
- PMID: 31506427
- PMCID: PMC6737147
- DOI: 10.1038/s41419-019-1874-9
Large-scale integrated analysis of ovarian cancer tumors and cell lines identifies an individualized gene expression signature for predicting response to platinum-based chemotherapy
Abstract
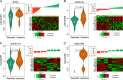
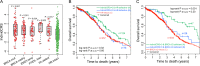
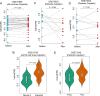
Heterogeneity in chemotherapeutic response is directly associated with prognosis and disease recurrence in patients with ovarian cancer (OvCa). Despite the significant clinical need, a credible gene signature for predicting response to platinum-based chemotherapy and for guiding the selection of personalized chemotherapy regimens has not yet been identified. The present study used an integrated approach involving both OvCa tumors and cell lines to identify an individualized gene expression signature, denoted as IndividCRS, consisting of 16 robust chemotherapy-responsive genes for predicting intrinsic or acquired chemotherapy response in the meta-discovery dataset. The robust performance of this signature was subsequently validated in 25 independent tumor datasets comprising 2215 patients and one independent cell line dataset, across different technical platforms. The IndividCRS was significantly correlated with the response to platinum therapy and predicted the improved outcome. Moreover, the IndividCRS correlated with homologous recombination deficiency (HRD) and was also capable of discriminating HR-deficient tumors with or without platinum-sensitivity for guiding HRD-targeted clinical trials. Our results reveal the universality and simplicity of the IndividCRS as a promising individualized genomic tool to rapidly monitor response to chemotherapy and predict the outcome of patients with OvCa.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
A Functional Homologous Recombination Assay Predicts Primary Chemotherapy Response and Long-Term Survival in Ovarian Cancer Patients.Clin Cancer Res. 2018 Sep 15;24(18):4482-4493. doi: 10.1158/1078-0432.CCR-17-3770. Epub 2018 Jun 1. Clin Cancer Res. 2018. PMID: 29858219
-
Systematic analysis reveals a lncRNA-mRNA co-expression network associated with platinum resistance in high-grade serous ovarian cancer.Invest New Drugs. 2018 Apr;36(2):187-194. doi: 10.1007/s10637-017-0523-3. Epub 2017 Oct 30. Invest New Drugs. 2018. PMID: 29082457
-
An integrated genomic-based approach to individualized treatment of patients with advanced-stage ovarian cancer.J Clin Oncol. 2007 Feb 10;25(5):517-25. doi: 10.1200/JCO.2006.06.3743. J Clin Oncol. 2007. Retraction in: J Clin Oncol. 2012 Feb 20;30(6):678. doi: 10.1200/jco.2012.42.0331. PMID: 17290060 Retracted.
-
Genomic and Epigenomic Signatures in Ovarian Cancer Associated with Resensitization to Platinum Drugs.Cancer Res. 2018 Feb 1;78(3):631-644. doi: 10.1158/0008-5472.CAN-17-1492. Epub 2017 Dec 11. Cancer Res. 2018. PMID: 29229600 Free PMC article.
-
Studying platinum sensitivity and resistance in high-grade serous ovarian cancer: Different models for different questions.Drug Resist Updat. 2016 Jan;24:55-69. doi: 10.1016/j.drup.2015.11.005. Epub 2015 Nov 26. Drug Resist Updat. 2016. PMID: 26830315 Review.
Cited by
-
Adaptive transcriptomic and immune infiltrate responses in the tumor immune microenvironment following neoadjuvant chemotherapy in high grade serous ovarian cancer reveal novel prognostic associations and activation of pro-tumorigenic pathways.Front Immunol. 2022 Sep 5;13:965331. doi: 10.3389/fimmu.2022.965331. eCollection 2022. Front Immunol. 2022. PMID: 36131935 Free PMC article.
-
TAF1A and ZBTB41 serve as novel key genes in cervical cancer identified by integrated approaches.Cancer Gene Ther. 2021 Dec;28(12):1298-1311. doi: 10.1038/s41417-020-00278-1. Epub 2020 Dec 12. Cancer Gene Ther. 2021. PMID: 33311601 Free PMC article.
-
Identification of the key genes associated with chemotherapy sensitivity in ovarian cancer patients.Cancer Med. 2020 Jul;9(14):5200-5209. doi: 10.1002/cam4.3122. Epub 2020 May 22. Cancer Med. 2020. PMID: 32441484 Free PMC article.
-
Gene networks and expression quantitative trait loci associated with adjuvant chemotherapy response in high-grade serous ovarian cancer.BMC Cancer. 2020 May 13;20(1):413. doi: 10.1186/s12885-020-06922-1. BMC Cancer. 2020. PMID: 32404140 Free PMC article.
-
Noncoding RNAs in gastric cancer: implications for drug resistance.Mol Cancer. 2020 Mar 19;19(1):62. doi: 10.1186/s12943-020-01185-7. Mol Cancer. 2020. PMID: 32192494 Free PMC article. Review.
References
-
- Doubeni CA, Doubeni AR, Myers AE. Diagnosis and management of ovarian cancer. Am. Fam. physician. 2016;93:937–944. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

