The CONSTANS flowering complex controls the protective response of photosynthesis in the green alga Chlamydomonas
- PMID: 31506429
- PMCID: PMC6736836
- DOI: 10.1038/s41467-019-11989-x
The CONSTANS flowering complex controls the protective response of photosynthesis in the green alga Chlamydomonas
Abstract
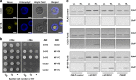
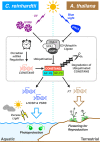
Light is essential for photosynthesis, but the amounts of light that exceed an organism's assimilation capacity can result in oxidative stress and even cell death. Plants and microalgae have developed a photoprotective response mechanism, qE, that dissipates excess light energy as thermal energy. In the green alga Chlamydomonas reinhardtii, qE is regulated by light-inducible photoprotective proteins, but the pathway from light perception to qE is not fully understood. Here, we show that the transcription factors CONSTANS and Nuclear transcription Factor Ys (NF-Ys) form a complex that governs light-dependent photoprotective responses in C. reinhardtii. The qE responses do not occur in CONSTANS or NF-Y mutants. The signal from light perception to the CONSTANS/NF-Ys complex is directly inhibited by the SPA1/COP1-dependent E3 ubiquitin ligase. This negative regulation mediated by the E3 ubiquitin ligase and the CONSTANS/NF-Ys complex is common to photoprotective response in algal photosynthesis and flowering in plants.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

