BioExcel Building Blocks, a software library for interoperable biomolecular simulation workflows
- PMID: 31506435
- PMCID: PMC6736963
- DOI: 10.1038/s41597-019-0177-4
BioExcel Building Blocks, a software library for interoperable biomolecular simulation workflows
Abstract
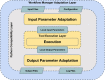
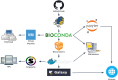
In the recent years, the improvement of software and hardware performance has made biomolecular simulations a mature tool for the study of biological processes. Simulation length and the size and complexity of the analyzed systems make simulations both complementary and compatible with other bioinformatics disciplines. However, the characteristics of the software packages used for simulation have prevented the adoption of the technologies accepted in other bioinformatics fields like automated deployment systems, workflow orchestration, or the use of software containers. We present here a comprehensive exercise to bring biomolecular simulations to the "bioinformatics way of working". The exercise has led to the development of the BioExcel Building Blocks (BioBB) library. BioBB's are built as Python wrappers to provide an interoperable architecture. BioBB's have been integrated in a chain of usual software management tools to generate data ontologies, documentation, installation packages, software containers and ways of integration with workflow managers, that make them usable in most computational environments.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
BioExcel Building Blocks REST API (BioBB REST API), programmatic access to interoperable biomolecular simulation tools.Bioinformatics. 2022 Jun 13;38(12):3302-3303. doi: 10.1093/bioinformatics/btac316. Bioinformatics. 2022. PMID: 35543460
-
BioExcel Building Blocks Workflows (BioBB-Wfs), an integrated web-based platform for biomolecular simulations.Nucleic Acids Res. 2022 Jul 5;50(W1):W99-W107. doi: 10.1093/nar/gkac380. Nucleic Acids Res. 2022. PMID: 35639735 Free PMC article.
-
Biowep: a workflow enactment portal for bioinformatics applications.BMC Bioinformatics. 2007 Mar 8;8 Suppl 1(Suppl 1):S19. doi: 10.1186/1471-2105-8-S1-S19. BMC Bioinformatics. 2007. PMID: 17430563 Free PMC article.
-
Development of an informatics infrastructure for data exchange of biomolecular simulations: Architecture, data models and ontology.SAR QSAR Environ Res. 2015;26(7-9):577-93. doi: 10.1080/1062936X.2015.1076515. Epub 2015 Sep 21. SAR QSAR Environ Res. 2015. PMID: 26387907 Free PMC article. Review.
-
Facilitating the use of large-scale biological data and tools in the era of translational bioinformatics.Brief Bioinform. 2014 Nov;15(6):942-52. doi: 10.1093/bib/bbt055. Epub 2013 Aug 1. Brief Bioinform. 2014. PMID: 23908249 Review.
Cited by
-
Conserved Water Networks Identification for Drug Design Using Density Clustering Approaches on Positional and Orientational Data.J Chem Inf Model. 2022 Dec 12;62(23):6105-6117. doi: 10.1021/acs.jcim.2c00801. Epub 2022 Nov 9. J Chem Inf Model. 2022. PMID: 36351288 Free PMC article.
-
BRIDGE: An Open Platform for Reproducible Protein-Ligand Simulations and Free Energy of Binding Calculations.Bio Protoc. 2020 Sep 5;10(17):e3731. doi: 10.21769/BioProtoc.3731. eCollection 2020 Sep 5. Bio Protoc. 2020. PMID: 33659392 Free PMC article.
-
Developments and applications of the OPTIMADE API for materials discovery, design, and data exchange.Digit Discov. 2024 Apr 18;3(8):1509-1533. doi: 10.1039/d4dd00039k. eCollection 2024 Aug 7. Digit Discov. 2024. PMID: 39118978 Free PMC article.
-
Intelligent Building Construction Management Based on BIM Digital Twin.Comput Intell Neurosci. 2021 Dec 14;2021:4979249. doi: 10.1155/2021/4979249. eCollection 2021. Comput Intell Neurosci. 2021. PMID: 34950199 Free PMC article.
-
pyKVFinder: an efficient and integrable Python package for biomolecular cavity detection and characterization in data science.BMC Bioinformatics. 2021 Dec 20;22(1):607. doi: 10.1186/s12859-021-04519-4. BMC Bioinformatics. 2021. PMID: 34930115 Free PMC article.
References
-
- Hospital A, Gelpi JL. High-throughput molecular dynamics simulations: toward a dynamic view of macromolecular structure. Wiley Interdisciplinary Reviews-Computational Molecular Science. 2013;3:364–377. doi: 10.1002/wcms.1142. - DOI

