Exosome-mediated uptake of mast cell tryptase into the nucleus of melanoma cells: a novel axis for regulating tumor cell proliferation and gene expression
- PMID: 31506436
- PMCID: PMC6736983
- DOI: 10.1038/s41419-019-1879-4
Exosome-mediated uptake of mast cell tryptase into the nucleus of melanoma cells: a novel axis for regulating tumor cell proliferation and gene expression
Abstract
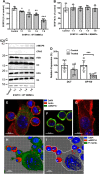
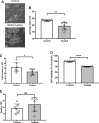
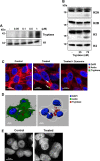
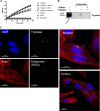
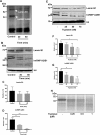
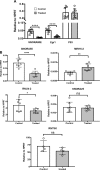
It is well established that mast cell accumulation accompanies most malignancies. However, the knowledge of how mast cells functionally impact on tumors is still rudimentary. Here we addressed this issue and show that mast cells have anti-proliferative activity on melanoma cells and that this effect is dependent on tryptase, a tetrameric protease stored in mast cell granules. Mechanistically, tryptase was found to be endocytosed by melanoma cells as cargo of DNA-coated exosomes released from melanoma cells, followed by transport to the nucleus. In the nucleus, tryptase executed clipping of histone 3 and degradation of Lamin B1, accompanied by extensive nuclear remodeling. Moreover, tryptase degraded hnRNP A2/B1, a protein involved in mRNA stabilization and interaction with non-coding RNAs. This was followed by downregulated expression of the oncogene EGR1 and of multiple non-coding RNAs, including oncogenic species. Altogether, these findings establish a new principle for regulation of tumor cell proliferation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Protective role of mouse mast cell tryptase Mcpt6 in melanoma.Pigment Cell Melanoma Res. 2020 Jul;33(4):579-590. doi: 10.1111/pcmr.12859. Epub 2020 Jan 19. Pigment Cell Melanoma Res. 2020. PMID: 31894627 Free PMC article.
-
The combined action of mast cell chymase, tryptase and carboxypeptidase A3 protects against melanoma colonization of the lung.Oncotarget. 2017 Apr 11;8(15):25066-25079. doi: 10.18632/oncotarget.15339. Oncotarget. 2017. PMID: 28212574 Free PMC article.
-
Tryptase-catalyzed core histone truncation: A novel epigenetic regulatory mechanism in mast cells.J Allergy Clin Immunol. 2017 Aug;140(2):474-485. doi: 10.1016/j.jaci.2016.11.044. Epub 2017 Jan 17. J Allergy Clin Immunol. 2017. PMID: 28108335
-
Mast cell tryptase - Marker and maker of cardiovascular diseases.Pharmacol Ther. 2019 Jul;199:91-110. doi: 10.1016/j.pharmthera.2019.03.008. Epub 2019 Mar 12. Pharmacol Ther. 2019. PMID: 30877022 Review.
-
The Role of Mast Cell Specific Chymases and Tryptases in Tumor Angiogenesis.Biomed Res Int. 2015;2015:142359. doi: 10.1155/2015/142359. Epub 2015 Jun 4. Biomed Res Int. 2015. PMID: 26146612 Free PMC article. Review.
Cited by
-
Signal Transduction Pathways Activated by Innate Immunity in Mast Cells: Translating Sensing of Changes into Specific Responses.Cells. 2020 Nov 4;9(11):2411. doi: 10.3390/cells9112411. Cells. 2020. PMID: 33158024 Free PMC article. Review.
-
Mast Cell: A Mysterious Character in Skin Cancer.In Vivo. 2024 Jan-Feb;38(1):58-68. doi: 10.21873/invivo.13410. In Vivo. 2024. PMID: 38148067 Free PMC article. Review.
-
Mast Cells and Dendritic Cells as Cellular Immune Checkpoints in Immunotherapy of Solid Tumors.Int J Mol Sci. 2022 Sep 21;23(19):11080. doi: 10.3390/ijms231911080. Int J Mol Sci. 2022. PMID: 36232398 Free PMC article. Review.
-
Controversial role of mast cells in NSCLC tumor progression and angiogenesis.Thorac Cancer. 2022 Nov;13(21):2929-2934. doi: 10.1111/1759-7714.14654. Epub 2022 Oct 4. Thorac Cancer. 2022. PMID: 36196487 Free PMC article. Review.
-
The Absence of Tryptase Mcpt6 Causes Elevated Cellular Stress in Response to Modulation of the Histone Acetylation Status in Mast Cells.Cells. 2019 Oct 2;8(10):1190. doi: 10.3390/cells8101190. Cells. 2019. PMID: 31581668 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

