Reduction of integrin alpha 4 activity through splice modulating antisense oligonucleotides
- PMID: 31506448
- PMCID: PMC6736852
- DOI: 10.1038/s41598-019-49385-6
Reduction of integrin alpha 4 activity through splice modulating antisense oligonucleotides
Abstract
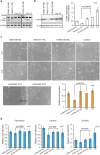
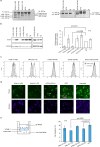
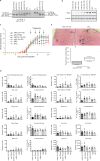
With recent approvals of antisense oligonucleotides as therapeutics, there is an increasing interest in expanding the application of these compounds to many other diseases. Our laboratory focuses on developing therapeutic splice modulating antisense oligonucleotides to treat diseases potentially amendable to intervention during pre-mRNA processing, and here we report the use of oligomers to down-regulate integrin alpha 4 protein levels. Over one hundred antisense oligonucleotides were designed to induce skipping of individual exons of the ITGA4 transcript and thereby reducing protein expression. Integrin alpha 4-mediated activities were evaluated in human dermal fibroblasts and Jurkat cells, an immortalised human T lymphocyte cell line. Peptide conjugated phosphorodiamidate morpholino antisense oligomers targeting ITGA4 were also assessed for their effect in delaying disease progression in the experimental autoimmune encephalomyelitis mouse model of multiple sclerosis. With the promising results in ameliorating disease progression, we are optimistic that the candidate oligomer may also be applicable to many other diseases associated with integrin alpha 4 mediated inflammation. This highly specific strategy to down-regulate protein expression through interfering with normal exon selection during pre-mRNA processing should be applicable to many other gene targets that undergo splicing during expression.
Conflict of interest statement
M.T.A.-H., S.F. and S.D.W. are named inventors of a patent describing the down-regulation of ITGA4: Multiple Sclerosis Treatment (20180104273), filed by Murdoch University, inventors: Stephen Donald Wilton, Sue Fletcher, May Aung-Htut, application status: pending. The
Figures

Similar articles
-
Translational development of splice-modifying antisense oligomers.Expert Opin Biol Ther. 2017 Jan;17(1):15-30. doi: 10.1080/14712598.2017.1250880. Epub 2016 Nov 2. Expert Opin Biol Ther. 2017. PMID: 27805416 Review.
-
Designing Effective Antisense Oligonucleotides for Exon Skipping.Methods Mol Biol. 2018;1687:143-155. doi: 10.1007/978-1-4939-7374-3_10. Methods Mol Biol. 2018. PMID: 29067661
-
Direct Reprogramming of Human DMD Fibroblasts into Myotubes for In Vitro Evaluation of Antisense-Mediated Exon Skipping and Exons 45-55 Skipping Accompanied by Rescue of Dystrophin Expression.Methods Mol Biol. 2018;1828:141-150. doi: 10.1007/978-1-4939-8651-4_8. Methods Mol Biol. 2018. PMID: 30171539 Free PMC article.
-
In Vitro Multiexon Skipping by Antisense PMOs in Dystrophic Dog and Exon 7-Deleted DMD Patient.Methods Mol Biol. 2018;1828:151-163. doi: 10.1007/978-1-4939-8651-4_9. Methods Mol Biol. 2018. PMID: 30171540 Free PMC article.
-
Recent Advances and Clinical Applications of Exon Inclusion for Spinal Muscular Atrophy.Methods Mol Biol. 2018;1828:57-68. doi: 10.1007/978-1-4939-8651-4_3. Methods Mol Biol. 2018. PMID: 30171534 Review.
Cited by
-
Antisense Oligonucleotide-Mediated Exon-skipping Therapies: Precision Medicine Spreading from Duchenne Muscular Dystrophy.JMA J. 2021 Jul 15;4(3):232-240. doi: 10.31662/jmaj.2021-0019. Epub 2021 Jul 9. JMA J. 2021. PMID: 34414317 Free PMC article. Review.
-
Developing predictive hybridization models for phosphorothioate oligonucleotides using high-resolution melting.PLoS One. 2022 May 18;17(5):e0268575. doi: 10.1371/journal.pone.0268575. eCollection 2022. PLoS One. 2022. PMID: 35584176 Free PMC article.
-
Polyglutamine Ataxias: Our Current Molecular Understanding and What the Future Holds for Antisense Therapies.Biomedicines. 2021 Oct 20;9(11):1499. doi: 10.3390/biomedicines9111499. Biomedicines. 2021. PMID: 34829728 Free PMC article. Review.
-
Dual Fluorescence Splicing Reporter Minigene Identifies an Antisense Oligonucleotide to Skip Exon v8 of the CD44 Gene.Int J Mol Sci. 2020 Nov 30;21(23):9136. doi: 10.3390/ijms21239136. Int J Mol Sci. 2020. PMID: 33266296 Free PMC article.
-
Expansion of Splice-Switching Therapy with Antisense Oligonucleotides.Int J Mol Sci. 2025 Mar 4;26(5):2270. doi: 10.3390/ijms26052270. Int J Mol Sci. 2025. PMID: 40076889 Free PMC article. Review.
References
-
- FDA grants accelerated approval to first drug for Duchenne muscular dystrophy (press release), https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm521263.htm (2016).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

