Genomic and developmental characterisation of a novel bunyavirus infecting the crustacean Carcinus maenas
- PMID: 31506463
- PMCID: PMC6736955
- DOI: 10.1038/s41598-019-49260-4
Genomic and developmental characterisation of a novel bunyavirus infecting the crustacean Carcinus maenas
Abstract
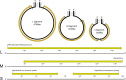
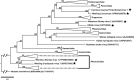
Carcinus maenas is in the top 100 globally invasive species and harbours a wide diversity of pathogens, including viruses. We provide a detailed description for a novel bunyavirus (Carcinus maenas Portunibunyavirus 1) infecting C. maenas from its native range in the Faroe Islands. The virus genome is tripartite, including large (L) (6766 bp), medium (M) (3244 bp) and small (S) (1608 bp) negative sense, single-stranded RNA segments. Individual genomic segments are flanked by 4 bp regions of similarity (CCUG). The segments encode an RNA-dependent RNA-polymerase, glycoprotein, non-structural protein with a Zinc-Finger domain and a nucleoprotein. Most show highest identity to the 'Wenling Crustacean Virus 9' from an unidentified crustacean host. Phylogenomics of crustacean-infecting bunyaviruses place them across multiple bunyavirus families. We discuss the diversity of crustacean bunyaviruses and provide an overview of how these viruses may affect the health and survival of crustacean hosts, including those inhabiting niches outside of their native range.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Molecular characterization of the African orthobunyavirus Ilesha virus.Infect Genet Evol. 2013 Dec;20:124-30. doi: 10.1016/j.meegid.2013.08.015. Epub 2013 Aug 26. Infect Genet Evol. 2013. PMID: 23988729
-
[Bunyavirus and its ecology].Uirusu. 2012;62(2):239-50. doi: 10.2222/jsv.62.239. Uirusu. 2012. PMID: 24153234 Review. Japanese.
-
Virus discovery in cultured portunid crabs: Genomic, phylogenetic, histopathological and microscopic characterization of a reovirus and a new bunyavirus.J Invertebr Pathol. 2024 Jun;204:108118. doi: 10.1016/j.jip.2024.108118. Epub 2024 Apr 27. J Invertebr Pathol. 2024. PMID: 38679369
-
Comparative characterization of the reassortant Orthobunyavirus Ngari with putative parental viruses, Bunyamwera and Batai: in vitro characterization and ex vivo stability.J Gen Virol. 2021 Feb;102(2):001523. doi: 10.1099/jgv.0.001523. J Gen Virol. 2021. PMID: 33258753 Free PMC article.
-
Viruses of the family Bunyaviridae: are all available isolates reassortants?Virology. 2013 Nov;446(1-2):207-16. doi: 10.1016/j.virol.2013.07.030. Epub 2013 Aug 30. Virology. 2013. PMID: 24074583 Review.
Cited by
-
A Novel Bunyavirus Discovered in Oriental Shrimp (Penaeus chinensis).Front Microbiol. 2021 Nov 24;12:751112. doi: 10.3389/fmicb.2021.751112. eCollection 2021. Front Microbiol. 2021. PMID: 34899637 Free PMC article.
-
Family Level Phylogenies Reveal Relationships of Plant Viruses within the Order Bunyavirales.Viruses. 2020 Sep 10;12(9):1010. doi: 10.3390/v12091010. Viruses. 2020. PMID: 32927652 Free PMC article.
-
Mycosis is a Disease State Encountered Rarely in Shore Crabs, Carcinus maenas.Pathogens. 2020 Jun 11;9(6):462. doi: 10.3390/pathogens9060462. Pathogens. 2020. PMID: 32545349 Free PMC article.
-
2021 Taxonomic update of phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales.Arch Virol. 2021 Dec;166(12):3513-3566. doi: 10.1007/s00705-021-05143-6. Arch Virol. 2021. PMID: 34463877 Free PMC article.
-
Enormous diversity of RNA viruses in economic crustaceans.mSystems. 2024 Oct 22;9(10):e0101624. doi: 10.1128/msystems.01016-24. Epub 2024 Sep 27. mSystems. 2024. PMID: 39329483 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

