Whole genome sequence analysis reveals high genetic variation of newly isolated Acidithiobacillus ferrooxidans IO-2C
- PMID: 31506467
- PMCID: PMC6736930
- DOI: 10.1038/s41598-019-49213-x
Whole genome sequence analysis reveals high genetic variation of newly isolated Acidithiobacillus ferrooxidans IO-2C
Abstract
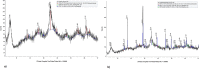
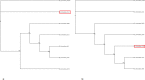
Acidithiobacillus ferrooxidans, a chemolithoautotrophic bacterium, is well known for its mineral oxidizing properties. The current study combines experimental and whole genome sequencing approaches to investigate an iron oxidizing, extreme acidophilic bacterium, A. ferrooxidans isolate (IO-2C) from an acid seep area near Carlos, TX, USA. Strain IO-2C was capable of oxidizing iron i.e. iron sulphate and iron ammonium sulphate yielding shwertmannite and jarosite minerals. Further, the bacterium's genome was sequenced, assembled and annotated to study its general features, structure and functions. To determine genetic heterogeneity, it was compared with the genomes of other published A. ferrooxidans strains. Pan-genome analysis displayed low gene conservation and significant genetic diversity in A. ferrooxidans species comprising of 6926 protein coding sequences with 23.04% (1596) core genes, 46.13% (3195) unique and 30.82% (2135) accessory genes. Variant analysis showed >75,000 variants, 287 of them with a predicted high impact, in A. ferrooxidans IO-2C genome compared to the reference strain, resulting in abandonment of some important functional key genes. The genome contains numerous functional genes for iron and sulphur metabolism, nitrogen fixation, secondary metabolites, degradation of aromatic compounds, and multidrug and heavy metal resistance. This study demonstrated the bio-oxidation of iron by newly isolated A. ferrooxidans IO-2C under acidic conditions, which was further supported by genomic analysis. Genomic analysis of this strain provided valuable information about the complement of genes responsible for the utilization of iron and tolerance of other metals.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Comparative genomics reveals intraspecific divergence of Acidithiobacillus ferrooxidans: insights from evolutionary adaptation.Microb Genom. 2023 Jun;9(6):mgen001038. doi: 10.1099/mgen.0.001038. Microb Genom. 2023. PMID: 37285209 Free PMC article.
-
Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans.BMC Genomics. 2009 Aug 24;10:394. doi: 10.1186/1471-2164-10-394. BMC Genomics. 2009. PMID: 19703284 Free PMC article.
-
Insights into the pathways of iron- and sulfur-oxidation, and biofilm formation from the chemolithotrophic acidophile Acidithiobacillus ferrivorans CF27.Res Microbiol. 2014 Nov;165(9):753-60. doi: 10.1016/j.resmic.2014.08.002. Epub 2014 Aug 19. Res Microbiol. 2014. PMID: 25154051
-
Genomic insights into the iron uptake mechanisms of the biomining microorganism Acidithiobacillus ferrooxidans.J Ind Microbiol Biotechnol. 2005 Dec;32(11-12):606-14. doi: 10.1007/s10295-005-0233-2. Epub 2005 May 14. J Ind Microbiol Biotechnol. 2005. PMID: 15895264 Review.
-
Acidithiobacillus ferrooxidans and its potential application.Extremophiles. 2018 Jul;22(4):563-579. doi: 10.1007/s00792-018-1024-9. Epub 2018 Apr 25. Extremophiles. 2018. PMID: 29696439 Review.
Cited by
-
Integrative Genomics Sheds Light on Evolutionary Forces Shaping the Acidithiobacillia Class Acidophilic Lifestyle.Front Microbiol. 2022 Feb 15;12:822229. doi: 10.3389/fmicb.2021.822229. eCollection 2021. Front Microbiol. 2022. PMID: 35242113 Free PMC article.
-
Genomic adaptations enabling Acidithiobacillus distribution across wide-ranging hot spring temperatures and pHs.Microbiome. 2021 Jun 11;9(1):135. doi: 10.1186/s40168-021-01090-1. Microbiome. 2021. PMID: 34116726 Free PMC article.
-
Draft Genome of Proteus mirabilis Serogroup O18 Elaborating Phosphocholine-Decorated O Antigen.Front Cell Infect Microbiol. 2021 Mar 25;11:620010. doi: 10.3389/fcimb.2021.620010. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33842384 Free PMC article.
-
Omics technology draws a comprehensive heavy metal resistance strategy in bacteria.World J Microbiol Biotechnol. 2024 May 6;40(6):193. doi: 10.1007/s11274-024-04005-y. World J Microbiol Biotechnol. 2024. PMID: 38709343 Review.
-
From Genes to Bioleaching: Unraveling Sulfur Metabolism in Acidithiobacillus Genus.Genes (Basel). 2023 Sep 8;14(9):1772. doi: 10.3390/genes14091772. Genes (Basel). 2023. PMID: 37761912 Free PMC article. Review.
References
-
- Sharma, A., Parashar, D. & Satyanarayana, T. Acidophilic microbes: biology and applications. 215–241 (Springer International Publishing, 2016).
-
- Babu, P., Chandel, A. K. & Singh, O. V. Survival mechanisms of extremophiles. 9–23 (Springer, 2015).
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

