Alginates along the filament of the brown alga Ectocarpus help cells cope with stress
- PMID: 31506545
- PMCID: PMC6736953
- DOI: 10.1038/s41598-019-49427-z
Alginates along the filament of the brown alga Ectocarpus help cells cope with stress
Abstract
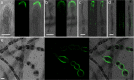
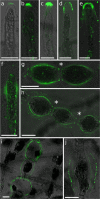
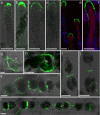
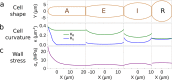
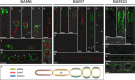
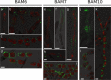
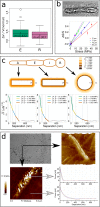
Ectocarpus is a filamentous brown alga, which cell wall is composed mainly of alginates and fucans (80%), two non-crystalline polysaccharide classes. Alginates are linear chains of epimers of 1,4-linked uronic acids, β-D-mannuronic acid (M) and α-L-guluronic acid (G). Previous physico-chemical studies showed that G-rich alginate gels are stiffer than M-rich alginate gels when prepared in vitro with calcium. In order to assess the possible role of alginates in Ectocarpus, we first immunolocalised M-rich or G-rich alginates using specific monoclonal antibodies along the filament. As a second step, we calculated the tensile stress experienced by the cell wall along the filament, and varied it with hypertonic or hypotonic solutions. As a third step, we measured the stiffness of the cell along the filament, using cell deformation measurements and atomic force microscopy. Overlapping of the three sets of data allowed to show that alginates co-localise with the stiffest and most stressed areas of the filament, namely the dome of the apical cell and the shanks of the central round cells. In addition, no major distinction between M-rich and G-rich alginate spatial patterns could be observed. Altogether, these results support that both M-rich and G-rich alginates play similar roles in stiffening the cell wall where the tensile stress is high and exposes cells to bursting, and that these roles are independent from cell growth and differentiation.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Baldauf SL. An overview of the phylogeny and diversity of eukaryotes. J. Syst. Evol. 2008;46:263–273.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

