Growth factor and receptor malfunctions associated with human genetic deafness
- PMID: 31506927
- PMCID: PMC8232520
- DOI: 10.1111/cge.13641
Growth factor and receptor malfunctions associated with human genetic deafness
Abstract
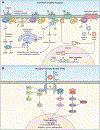
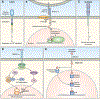
A variety of different signaling pathways are necessary for development and maintenance of the human auditory system. Normal hearing allows for the detection of soft sounds within the frequency range of 20 to 20 000 Hz, but more importantly to perceive the human voice frequency band of 250 to 6000 Hz. Loss of hearing is common, and is a clinically heterogeneous disorder that can be caused by environmental factors such as exposure to loud noise, infections and ototoxic drugs. In addition, variants of hundreds of genes have been reported to disrupt processes required for hearing. Noncoding regulatory variants and variants of additional genes necessary for hearing remain to be discovered as many individuals with inherited deafness are without a genetic diagnosis, despite the advent of whole exome sequencing. Here, we discuss in detail some of these deafness-causing variants of genes encoding a ligand or its receptor. Spotlighted in this review are three growth factor-receptor-pairs EDN3/EDNRB, HGF/MET and JAG/NOTCH, which individually are necessary for normal hearing. We also offer our perspective on unanswered questions, future challenges and potential opportunities for treatments emerging from molecular genetic and mechanistic studies of deafness due to these causes.
Keywords: EDN3; EDNRB; HGF; JAG; MET; NOTCH; hearing; inner ear defects.
© 2019 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Conflict of interest statement
DISCLOSURE
The authors declare no conflict of interest
Figures

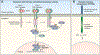
References
-
- Tucci D, Merson MH, Wilson BS. A summary of the literature on global hearing impairment: current status and priorities for action. Otol Neurotol. 2010;31(1):31–41. - PubMed
-
- Yueh B, Shapiro N, MacLean CH, Shekelle PG. Screening and management of adult hearing loss in primary care: scientific review. JAMA. 2003;289(15):1976–1985. - PubMed
-
- Purves D, Augustine GJ, Fitzpatrick D, et al., eds. Neuroscience. 2 ed: Sunderland; 2001.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

