Insights From Molecular Dynamics Simulations of a Number of G-Protein Coupled Receptor Targets for the Treatment of Pain and Opioid Use Disorders
- PMID: 31507375
- PMCID: PMC6716474
- DOI: 10.3389/fnmol.2019.00207
Insights From Molecular Dynamics Simulations of a Number of G-Protein Coupled Receptor Targets for the Treatment of Pain and Opioid Use Disorders
Abstract
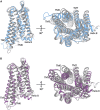
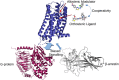
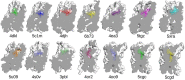
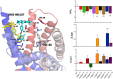
Effective treatments for pain management remain elusive due to the dangerous side-effects of current gold-standard opioid analgesics, including the respiratory depression that has led to skyrocketing death rates from opioid overdoses over the past decade. In an attempt to address the horrific opioid crisis worldwide, the National Institute on Drug Abuse has recently proposed boosting research on specific pharmacological mechanisms mediated by a number of G protein-coupled receptors (GPCRs). This research is expected to expedite the discovery of medications for opioid overdose and opioid use disorders, leading toward a safer and more effective treatment of pain. Here, we review mechanistic insights from recent all-atom molecular dynamics simulations of a specific subset of GPCRs for which high-resolution experimental structures are available, including opioid, cannabinoid, orexin, metabotropic glutamate, and dopamine receptor subtypes.
Keywords: GPCRs; molecular dynamics; opioid crisis; opioid use disorder; pain.
Figures
References
-
- An X., Bai Q., Bing Z., Zhou S., Shi D., Liu H., et al. (2018). How does agonist and antagonist binding lead to different conformational ensemble equilibria of the κ-Opioid receptor: insight from long-time gaussian accelerated molecular dynamics simulation. ACS Chem. Neurosci. 10 1575–1584. 10.1021/acschemneuro.8b00535 - DOI - PubMed
-
- Ballesteros J. A., Weinstein H. (1995). “Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors,” in Methods in Neurosciences, eds Sealfon S. C., Conn P. M., (San Diego, CA: Academic Press; ), 366–428. 10.1016/s1043-9471(05)80049-7 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

