Genomic and Proteomic Characterizations of Sfin-1, a Novel Lytic Phage Infecting Multidrug-Resistant Shigella spp. and Escherichia coli C
- PMID: 31507544
- PMCID: PMC6714547
- DOI: 10.3389/fmicb.2019.01876
Genomic and Proteomic Characterizations of Sfin-1, a Novel Lytic Phage Infecting Multidrug-Resistant Shigella spp. and Escherichia coli C
Abstract
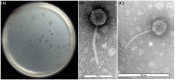
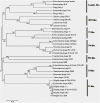
Shigellosis is a public health threat in developed as well as developing countries like "India." While antibiotic therapy is the mainstay of treatment for shigellosis, current emergence of multidrug-resistant strains of Shigella spp. has posed the problem more challenging. Lytic bacteriophages which destroy antibiotic resistant Shigella spp. have great potential in this context and hence their identification and detailed characterization is necessary. In this study we presented the isolation and a detailed characterization of a novel bacteriophage Sfin-1, which shows potent lytic activity against multidrug-resistant isolates of Shigella flexneri, Shigella dysenteriae, Shigella sonnei obtained from clinical specimens from shigellosis patients. It is also active against Escherichia coli C. The purified phage is lytic in nature, exhibited absorption within 5-10 min, a latent period of 5-20 min and burst size of ∼28 to ∼146 PFU/cell. The isolated phage shows stability in a broad pH range and survives an hour at 50°C. Genome sequencing and phylogenetic analyses showed that Sfin-1 is a novel bacteriophage, which is very closely related to T1-like phages (89.59% identity with Escherichia virus T1). In silico analysis indicates that Sfin-1 genome consists of double stranded linear DNA of 50,403 bp (GC content of 45.2%) encoding 82 potential coding sequences, several potential promoters and transcriptional terminators. Under electron microscopy, Sfin-1 shows morphology characteristics of the family Siphoviridae with an isometric head (61 nm) and a non-contractile tail (155 nm). This is most likely the first report of a lytic bacteriophage that is active against three of the most virulent multidrug-resistant Shigella species and therefore might have a potential role in phage therapy of patients infected with these organisms.
Keywords: LC-MS/MS; Shigella spp.; bacteriophage; genome sequencing; large terminase; phage therapy.
Figures

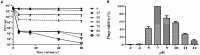


Similar articles
-
Characterizations of novel broad-spectrum lytic bacteriophages Sfin-2 and Sfin-6 infecting MDR Shigella spp. with their application on raw chicken to reduce the Shigella load.Front Microbiol. 2023 Nov 29;14:1240570. doi: 10.3389/fmicb.2023.1240570. eCollection 2023. Front Microbiol. 2023. PMID: 38094623 Free PMC article.
-
Isolation, characterization and genomic analysis of a novel lytic bacteriophage vB_SsoS-ISF002 infecting Shigella sonnei and Shigella flexneri.J Med Microbiol. 2018 Mar;67(3):376-386. doi: 10.1099/jmm.0.000683. Epub 2018 Jan 30. J Med Microbiol. 2018. PMID: 29458535
-
Morphological, biological, and genomic characterization of a newly isolated lytic phage Sfk20 infecting Shigella flexneri, Shigella sonnei, and Shigella dysenteriae1.Sci Rep. 2021 Sep 29;11(1):19313. doi: 10.1038/s41598-021-98910-z. Sci Rep. 2021. PMID: 34588569 Free PMC article.
-
Characterization and complete genome sequence of the Shigella bacteriophage pSf-1.Res Microbiol. 2013 Dec;164(10):979-86. doi: 10.1016/j.resmic.2013.08.007. Epub 2013 Sep 5. Res Microbiol. 2013. PMID: 24012542
-
Shigellosis.J Microbiol. 2005 Apr;43(2):133-43. J Microbiol. 2005. PMID: 15880088 Review.
Cited by
-
Ecology, Structure, and Evolution of Shigella Phages.Annu Rev Virol. 2020 Sep 29;7(1):121-141. doi: 10.1146/annurev-virology-010320-052547. Epub 2020 May 11. Annu Rev Virol. 2020. PMID: 32392456 Free PMC article. Review.
-
Isolation and characterization of two homolog phages infecting Pseudomonas aeruginosa.Front Microbiol. 2022 Jul 14;13:946251. doi: 10.3389/fmicb.2022.946251. eCollection 2022. Front Microbiol. 2022. PMID: 35935197 Free PMC article.
-
Potential for Phages in the Treatment of Bacterial Sexually Transmitted Infections.Antibiotics (Basel). 2021 Aug 24;10(9):1030. doi: 10.3390/antibiotics10091030. Antibiotics (Basel). 2021. PMID: 34572612 Free PMC article. Review.
-
Proteomic Analysis of Marine Bacteriophages: Structural Conservation, Post-Translational Modifications, and Phage-Host Interactions.Environ Microbiol. 2025 Apr;27(4):e70099. doi: 10.1111/1462-2920.70099. Environ Microbiol. 2025. PMID: 40262907 Free PMC article.
-
Characterization and comparative genomic analysis of novel lytic bacteriophages targeting Cronobacter sakazakii.Virus Res. 2023 May;329:199102. doi: 10.1016/j.virusres.2023.199102. Epub 2023 Apr 1. Virus Res. 2023. PMID: 36963724 Free PMC article.
References
-
- Amarillas L., Rubi-Rangel L., Chaidez C., Gonzalez-Robles A., Lightbourn-Rojas L., Leon-Felix J. (2017). Isolation and characterization of phiLLS, a novel phage with potential biocontrol agent against multidrug-resistant Escherichia coli. Front. Microbiol. 8:1355. 10.3389/fmicb.2017.01355 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

