New Halonotius Species Provide Genomics-Based Insights Into Cobalamin Synthesis in Haloarchaea
- PMID: 31507553
- PMCID: PMC6719526
- DOI: 10.3389/fmicb.2019.01928
New Halonotius Species Provide Genomics-Based Insights Into Cobalamin Synthesis in Haloarchaea
Abstract
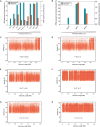
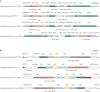
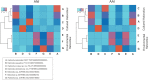
Hypersaline aquatic and terrestrial ecosystems display a cosmopolitan distribution. These environments teem with microbes and harbor a plethora of prokaryotic lineages that evaded ecological characterization due to the prior inability to cultivate them or to access their genomic information. In order to close the current knowledge gap, we performed two sampling and isolation campaigns in the saline soils of the Odiel Saltmarshes and the salterns of Isla Cristina (Huelva, Spain). From the isolated haloarchaeal strains subjected to high-throughput phylogenetic screening, two were chosen (F15BT and F9-27T) for physiological and genomic characterization due of their relatedness to the genus Halonotius. Comparative genomic analyses were carried out between the isolated strains and the genomes of previously described species Halonotius pteroides CECT 7525T, Halonotius aquaticus F13-13T and environmentaly recovered metagenome-assembled representatives of the genus Halonotius. The topology of the phylogenomic tree showed agreement with the phylogenetic ones based on 16S rRNA and rpoB' genes, and together with average amino acid and nucleotide identities suggested the two strains as novel species within the genus. We propose the names Halonotius terrestris sp. nov. (type strain F15BT = CECT 9688T = CCM 8954T) and Halonotius roseus sp. nov. (type strain F9-27T = CECT 9745T = CCM 8956T) for these strains. Comparative genomic analyses within the genus highlighted a typical salt-in signature, characterized by acidic proteomes with low isoelectric points, and indicated heterotrophic aerobic lifestyles. Genome-scale metabolic reconstructions revealed that the newly proposed species encode all the necessary enzymatic reactions involved in cobalamin (vitamin B12) biosynthesis. Based on the worldwide distribution of the genus and its abundance in hypersaline habitats we postulate that its members perform a critical function by being able to provide "expensive" commodities (i.e., vitamin B12) to the halophilic microbial communities at large.
Keywords: Halonotius; Halonotius roseus sp. nov.; Halonotius terrestris sp. nov.; comparative genomic analysis; haloarchaea; hypersaline environment.
Figures





References
Associated data
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

