Functional Genomic Analyses of Exopolysaccharide-Producing Streptococcus thermophilus ASCC 1275 in Response to Milk Fermentation Conditions
- PMID: 31507577
- PMCID: PMC6716118
- DOI: 10.3389/fmicb.2019.01975
Functional Genomic Analyses of Exopolysaccharide-Producing Streptococcus thermophilus ASCC 1275 in Response to Milk Fermentation Conditions
Abstract
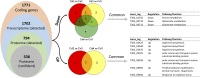
Exopolysaccharide (EPS) produced from dairy bacteria improves texture and functionalities of fermented dairy foods. Our previous study showed improved EPS production from Streptococcus thermophilus ASCC1275 (ST1275) by simple alteration of fermentation conditions such as pH decrease (pH 6.5 → pH 5.5), temperature increase (37°C → 40°C) and/or whey protein isolate (WPI) supplementation. The iTRAQ-based proteomics in combination with transcriptomics were applied to understand cellular protein expression in ST1275 in response to above shifts during milk fermentation. The pH decrease induced the most differentially expressed proteins (DEPs) that are involved in cellular metabolic responses including glutamate catabolism, arginine biosynthesis, cysteine catabolism, purine metabolism, lactose uptake, and fatty acid biosynthesis. Temperature increase and WPI supplementation did not induce much changes in global protein express profiles of ST1275 between comparisons of pH 5.5 conditions. Comparative proteomic analyses from pairwise comparisons demonstrated enhanced glutamate catabolism and purine metabolism under pH 5.5 conditions (Cd2, Cd3, and Cd4) compared to that of pH 6.5 condition (Cd1). Concordance analysis for differential expressed genes (DEGs) and DEPs highlighted down-regulated glutamate catabolism and up-regulated arginine biosynthesis in pH 5.5 conditions. Down regulation of glutamate catabolism was also confirmed by pathway enrichment analysis. Down-regulation of EpsB involved in EPS assembly was observed at both mRNA and protein level in pH 5.5 conditions compared to that in pH 6.5 condition. Medium pH decreased to mild acidic level induced cellular changes associated with glutamate catabolism, arginine biosynthesis and regulation of EPS assembly in ST1275.
Keywords: Streptococcus thermophilus; exopolysaccharide; pH; proteome; transcriptome.
Figures





Similar articles
-
Comparative mRNA-Seq Analysis Reveals the Improved EPS Production Machinery in Streptococcus thermophilus ASCC 1275 During Optimized Milk Fermentation.Front Microbiol. 2018 Mar 13;9:445. doi: 10.3389/fmicb.2018.00445. eCollection 2018. Front Microbiol. 2018. PMID: 29593689 Free PMC article.
-
Fermentation conditions affecting the bacterial growth and exopolysaccharide production by Streptococcus thermophilus ST 111 in milk-based medium.J Appl Microbiol. 2004;97(6):1257-73. doi: 10.1111/j.1365-2672.2004.02418.x. J Appl Microbiol. 2004. PMID: 15546417
-
Effects of pH, temperature, supplementation with whey protein concentrate, and adjunct cultures on the production of exopolysaccharides by Streptococcus thermophilus 1275.J Dairy Sci. 2003 Nov;86(11):3405-15. doi: 10.3168/jds.S0022-0302(03)73944-7. J Dairy Sci. 2003. PMID: 14672169
-
Biochemistry, genetics, and applications of exopolysaccharide production in Streptococcus thermophilus: a review.J Dairy Sci. 2003 Feb;86(2):407-23. doi: 10.3168/jds.S0022-0302(03)73619-4. J Dairy Sci. 2003. PMID: 12647947 Review.
-
ADSA Foundation Scholar Award: Possibilities and challenges of exopolysaccharide-producing lactic cultures in dairy foods.J Dairy Sci. 2008 Apr;91(4):1282-98. doi: 10.3168/jds.2007-0558. J Dairy Sci. 2008. PMID: 18349221 Review.
Cited by
-
Advances in fermented foods revealed by multi-omics: A new direction toward precisely clarifying the roles of microorganisms.Front Microbiol. 2022 Dec 14;13:1044820. doi: 10.3389/fmicb.2022.1044820. eCollection 2022. Front Microbiol. 2022. PMID: 36590428 Free PMC article. Review.
-
Exopolysaccharides From Streptococcus thermophilus ST538 Modulate the Antiviral Innate Immune Response in Porcine Intestinal Epitheliocytes.Front Microbiol. 2020 May 19;11:894. doi: 10.3389/fmicb.2020.00894. eCollection 2020. Front Microbiol. 2020. PMID: 32508770 Free PMC article.
-
Genome-Scale Metabolic Modeling Combined with Transcriptome Profiling Provides Mechanistic Understanding of Streptococcus thermophilus CH8 Metabolism.Appl Environ Microbiol. 2022 Aug 23;88(16):e0078022. doi: 10.1128/aem.00780-22. Epub 2022 Aug 4. Appl Environ Microbiol. 2022. PMID: 35924931 Free PMC article.
-
Use of Cell Envelope Targeting Antibiotics and Antimicrobial Agents as a Powerful Tool to Select for Lactic Acid Bacteria Strains With Improved Texturizing Ability in Milk Fermentations.Front Bioeng Biotechnol. 2021 Jan 13;8:623700. doi: 10.3389/fbioe.2020.623700. eCollection 2020. Front Bioeng Biotechnol. 2021. PMID: 33520973 Free PMC article.
-
Structure Characterization of Polysaccharide from Chinese Yam (Dioscorea opposite Thunb.) and Its Growth-Promoting Effects on Streptococcus thermophilus.Foods. 2021 Nov 4;10(11):2698. doi: 10.3390/foods10112698. Foods. 2021. PMID: 34828979 Free PMC article.
References
-
- Amatayakul T., Halmos A. L., Sherkat F., Shah N. P. (2006a). Physical characteristics of yoghurts made using exopolysaccharide-producing starter cultures and varying casein to whey protein ratios. Int. Dairy J. 16 40–51. 10.1016/j.idairyj.2005.01.004 - DOI
-
- Amatayakul T., Sherkat F., Shah N. P. (2006b). Physical characteristics of set yoghurt made with altered casein to whey protein ratios and EPS-producing starter cultures at 9 and 14% total solids. Food Hydrocoll. 20 314–324. 10.1016/j.foodhyd.2005.02.015 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

