MEK1/2 Inhibitors Unlock the Constrained Interferon Response in Macrophages Through IRF1 Signaling
- PMID: 31507609
- PMCID: PMC6718554
- DOI: 10.3389/fimmu.2019.02020
MEK1/2 Inhibitors Unlock the Constrained Interferon Response in Macrophages Through IRF1 Signaling
Abstract
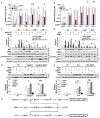
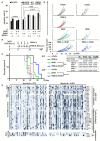
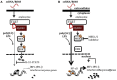
Macrophages are immune sentinels essential for pathogen recognition and immune defense. Nucleic acid-sensing toll-like receptors like TLR7 activate tailored proinflammatory and interferon responses in macrophages. Here we found that TLR7 activation constrained itself and other TLRs from inducing interferon response genes in macrophages through MAPK kinase 1/2 (MEK1/2)-dependent IRF1 inhibition. Downstream of the MEK1/2-ERK pathway, TLR7-activated macrophages induced interleukin-10 (IL-10), a signal transducer and activator of transcription 3 (STAT3) signaling axis, which constrained the expression of interferon response genes, immunomodulatory cytokines, and chemokines. Nevertheless, MEK1/2 inhibitors unlocked an IRF1-interferon signature response in an NF-κB-dependent manner. Deficiency in interferon regulatory factor 1 (Irf1) completely abrogated the interferon response and prevented the reprogramming of macrophages into an immunostimulatory phenotype. As a proof of concept, combination treatment with a TLR7 agonist and MEK1/2 inhibitor synergistically extended the survival of wild-type but not Irf1-deficient melanoma-bearing mice. In a retrospective study, higher expression of Irf1 and interferon response genes correlated with more favorable prognosis in patients with cutaneous melanoma. Our findings demonstrated how MEK1/2 inhibitor unlocks IRF1-mediated interferon signature response in macrophages, and the therapeutic potentials of combination therapy with MEK1/2 inhibitor and TLR7 agonist.
Keywords: IRF1; MAPK; MEK inhibitor; TLR7 agonist; interferon; macrophage; reprogramming.
Figures





Similar articles
-
The MEK1/2-ERK Pathway Inhibits Type I IFN Production in Plasmacytoid Dendritic Cells.Front Immunol. 2018 Feb 26;9:364. doi: 10.3389/fimmu.2018.00364. eCollection 2018. Front Immunol. 2018. PMID: 29535732 Free PMC article.
-
Restriction of Legionella pneumophila replication in macrophages requires concerted action of the transcriptional regulators Irf1 and Irf8 and nod-like receptors Naip5 and Nlrc4.Infect Immun. 2009 Nov;77(11):4794-805. doi: 10.1128/IAI.01546-08. Epub 2009 Aug 31. Infect Immun. 2009. PMID: 19720760 Free PMC article.
-
Inhibitory effect of 10-hydroxydecanoic acid on lipopolysaccharide-induced nitric oxide production via translational downregulation of interferon regulatory factor-1 in RAW264 murine macrophages.Biomed Res. 2013 Aug;34(4):205-14. doi: 10.2220/biomedres.34.205. Biomed Res. 2013. PMID: 23995057
-
A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses.Front Immunol. 2018 May 28;9:1135. doi: 10.3389/fimmu.2018.01135. eCollection 2018. Front Immunol. 2018. PMID: 29892288 Free PMC article. Review.
-
Regulation of interferon and Toll-like receptor signaling during macrophage activation by opposing feedforward and feedback inhibition mechanisms.Immunol Rev. 2008 Dec;226:41-56. doi: 10.1111/j.1600-065X.2008.00707.x. Immunol Rev. 2008. PMID: 19161415 Free PMC article. Review.
Cited by
-
MAP kinase kinase 1 (MEK1) within extracellular vesicles inhibits tumour growth by promoting anti-tumour immunity.J Extracell Vesicles. 2024 Oct;13(10):e12515. doi: 10.1002/jev2.12515. J Extracell Vesicles. 2024. PMID: 39330930 Free PMC article.
-
mtSTAT3 suppresses rheumatoid arthritis by regulating Th17 and synovial fibroblast inflammatory cell death with IL-17-mediated autophagy dysfunction.Exp Mol Med. 2025 Feb;57(1):221-234. doi: 10.1038/s12276-024-01376-y. Epub 2025 Jan 17. Exp Mol Med. 2025. PMID: 39825104 Free PMC article.
-
EGFR Activation Impairs Antiviral Activity of Interferon Signaling in Brain Microvascular Endothelial Cells During Japanese Encephalitis Virus Infection.Front Microbiol. 2022 Jun 30;13:894356. doi: 10.3389/fmicb.2022.894356. eCollection 2022. Front Microbiol. 2022. PMID: 35847084 Free PMC article.
-
Systems Immunology Analysis Reveals an Immunomodulatory Effect of Snail-p53 Binding on Neutrophil- and T Cell-Mediated Immunity in KRAS Mutant Non-Small Cell Lung Cancer.Front Immunol. 2020 Dec 14;11:569671. doi: 10.3389/fimmu.2020.569671. eCollection 2020. Front Immunol. 2020. PMID: 33381110 Free PMC article.
-
Intertumoral heterogeneity impacts oncolytic vesicular stomatitis virus efficacy in mouse pancreatic cancer cells.J Virol. 2023 Sep 28;97(9):e0100523. doi: 10.1128/jvi.01005-23. Epub 2023 Sep 6. J Virol. 2023. PMID: 37671865 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

