Identification and manipulation of Neurospora crassa genes involved in sensitivity to furfural
- PMID: 31508149
- PMCID: PMC6724289
- DOI: 10.1186/s13068-019-1550-4
Identification and manipulation of Neurospora crassa genes involved in sensitivity to furfural
Abstract
Background: Biofuels derived from lignocellulosic biomass are a viable alternative to fossil fuels required for transportation. Following plant biomass pretreatment, the furan derivative furfural is present at concentrations which are inhibitory to yeasts. Detoxification of furfural is thus important for efficient fermentation. Here, we searched for new genetic attributes in the fungus Neurospora crassa that may be linked to furfural tolerance. The fact that furfural is involved in the natural process of sexual spore germination of N. crassa and that this fungus is highly amenable to genetic manipulations makes it a rational candidate for this study.
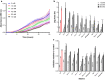
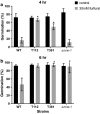
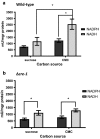
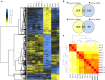
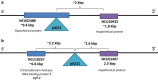
Results: Both hypothesis-based and unbiased (random promotor mutagenesis) approaches were performed to identify N. crassa genes associated with the response to furfural. Changes in the transcriptional profile following exposure to furfural revealed that the affected processes were, overall, similar to those observed in Saccharomyces cerevisiae. N. crassa was more tolerant (by ~ 30%) to furfural when carboxymethyl cellulose was the main carbon source as opposed to sucrose, indicative of a link between carbohydrate metabolism and furfural tolerance. We also observed increased tolerance in a Δcre-1 mutant (CRE-1 is a key transcription factor that regulates the ability of fungi to utilize non-preferred carbon sources). In addition, analysis of aldehyde dehydrogenase mutants showed that ahd-2 (NCU00378) was involved in tolerance to furfural as well as the predicted membrane transporter NCU05580 (flr-1), a homolog of FLR1 in S. cerevisiae. Further to the rational screening, an unbiased approach revealed additional genes whose inactivation conferred increased tolerance to furfural: (i) NCU02488, which affected the abundance of the non-anchored cell wall protein NCW-1 (NCU05137), and (ii) the zinc finger protein NCU01407.
Conclusions: We identified attributes in N. crassa associated with tolerance or degradation of furfural, using complementary research approaches. The manipulation of the genes involved in furan sensitivity can provide a means for improving the production of biofuel producing strains. Similar research approaches can be utilized in N. crassa and other filamentous fungi to identify additional attributes relevant to other furans or toxic chemicals.
Keywords: Aldehyde dehydrogenases; CRE1; Furan; Furfural; Neurospora crassa; Pretreatment.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures
References
-
- Zheng Y, Shi J, Tu M, Cheng Y-S. Principles and development of lignocellulosic biomass pretreatment for biofuels. In: Advances in bioenergy. Elsevier; 2017. p. 1–68.
-
- Bhutto AW, Qureshi K, Harijan K, Abro R, Abbas T, Bazmi AA, et al. Insight into progress in pre-treatment of lignocellulosic biomass. Energy. 2017;122:724–745. doi: 10.1016/j.energy.2017.01.005. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

