Large-scale sequence comparisons with sourmash
- PMID: 31508216
- PMCID: PMC6720031
- DOI: 10.12688/f1000research.19675.1
Large-scale sequence comparisons with sourmash
Abstract
The sourmash software package uses MinHash-based sketching to create "signatures", compressed representations of DNA, RNA, and protein sequences, that can be stored, searched, explored, and taxonomically annotated. sourmash signatures can be used to estimate sequence similarity between very large data sets quickly and in low memory, and can be used to search large databases of genomes for matches to query genomes and metagenomes. sourmash is implemented in C++, Rust, and Python, and is freely available under the BSD license at http://github.com/dib-lab/sourmash.
Keywords: MinHash; bioinformatics; k-mer; sequence analysis; sourmash.
Conflict of interest statement
No competing interests were disclosed.
Figures
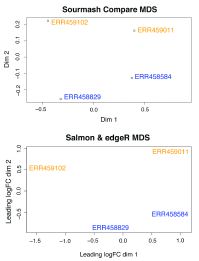


References
-
- Sequence read archive overview.2018. Reference Source
-
- Broder AZ: On the resemblance and containment of documents. In Compression and complexity of sequences 1997. proceedings.IEEE.1997;21–29. Reference Source
-
- Bovee R, Greenfield N: Finch: a tool adding dynamic abundance filtering to genomic minhashing.2018;3(22):505 10.21105/joss.00505 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

