Staining Invariant Features for Improving Generalization of Deep Convolutional Neural Networks in Computational Pathology
- PMID: 31508414
- PMCID: PMC6716536
- DOI: 10.3389/fbioe.2019.00198
Staining Invariant Features for Improving Generalization of Deep Convolutional Neural Networks in Computational Pathology
Abstract
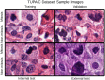
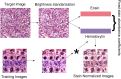
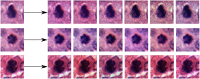
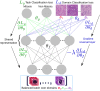
One of the main obstacles for the implementation of deep convolutional neural networks (DCNNs) in the clinical pathology workflow is their low capability to overcome variability in slide preparation and scanner configuration, that leads to changes in tissue appearance. Some of these variations may not be not included in the training data, which means that the models have a risk to not generalize well. Addressing such variations and evaluating them in reproducible scenarios allows understanding of when the models generalize better, which is crucial for performance improvements and better DCNN models. Staining normalization techniques (often based on color deconvolution and deep learning) and color augmentation approaches have shown improvements in the generalization of the classification tasks for several tissue types. Domain-invariant training of DCNN's is also a promising technique to address the problem of training a single model for different domains, since it includes the source domain information to guide the training toward domain-invariant features, achieving state-of-the-art results in classification tasks. In this article, deep domain adaptation in convolutional networks (DANN) is applied to computational pathology and compared with widely used staining normalization and color augmentation methods in two challenging classification tasks. The classification tasks rely on two openly accessible datasets, targeting Gleason grading in prostate cancer, and mitosis classification in breast tissue. The benchmark of the different techniques and their combination in two DCNN architectures allows us to assess the generalization abilities and advantages of each method in the considered classification tasks. The code for reproducing our experiments and preprocessing the data is publicly available. Quantitative and qualitative results show that the use of DANN helps model generalization to external datasets. The combination of several techniques to manage color heterogeneity suggests that several methods together, such as color augmentation methods with DANN training, can generalize even further. The results do not show a single best technique among the considered methods, even when combining them. However, color augmentation and DANN training obtain most often the best results (alone or combined with color normalization and color augmentation). The statistical significance of the results and the embeddings visualizations provide useful insights to design DCNN that generalizes to unseen staining appearances. Furthermore, in this work, we release for the first time code for DANN evaluation in open access datasets for computational pathology. This work opens the possibility for further research on using DANN models together with techniques that can overcome the tissue preparation differences across datasets to tackle limited generalization.
Keywords: adversarial neural networks; color augmentation; color normalization; digital pathology; domain shift; staining normalization.
Figures



Similar articles
-
Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology.Med Image Anal. 2019 Dec;58:101544. doi: 10.1016/j.media.2019.101544. Epub 2019 Aug 21. Med Image Anal. 2019. PMID: 31466046
-
The role of unpaired image-to-image translation for stain color normalization in colorectal cancer histology classification.Comput Methods Programs Biomed. 2023 Jun;234:107511. doi: 10.1016/j.cmpb.2023.107511. Epub 2023 Mar 26. Comput Methods Programs Biomed. 2023. PMID: 37011426
-
Semi-supervised training of deep convolutional neural networks with heterogeneous data and few local annotations: An experiment on prostate histopathology image classification.Med Image Anal. 2021 Oct;73:102165. doi: 10.1016/j.media.2021.102165. Epub 2021 Jul 14. Med Image Anal. 2021. PMID: 34303169
-
Generative Adversarial Networks in Digital Pathology and Histopathological Image Processing: A Review.J Pathol Inform. 2021 Nov 3;12:43. doi: 10.4103/jpi.jpi_103_20. eCollection 2021. J Pathol Inform. 2021. PMID: 34881098 Free PMC article. Review.
-
Deep learning for electroencephalogram (EEG) classification tasks: a review.J Neural Eng. 2019 Jun;16(3):031001. doi: 10.1088/1741-2552/ab0ab5. Epub 2019 Feb 26. J Neural Eng. 2019. PMID: 30808014 Review.
Cited by
-
Combining weakly and strongly supervised learning improves strong supervision in Gleason pattern classification.BMC Med Imaging. 2021 May 8;21(1):77. doi: 10.1186/s12880-021-00609-0. BMC Med Imaging. 2021. PMID: 33964886 Free PMC article.
-
Learning generalizable AI models for multi-center histopathology image classification.NPJ Precis Oncol. 2024 Jul 19;8(1):151. doi: 10.1038/s41698-024-00652-4. NPJ Precis Oncol. 2024. PMID: 39030380 Free PMC article.
-
CNN stability training improves robustness to scanner and IHC-based image variability for epithelium segmentation in cervical histology.Front Med (Lausanne). 2023 Jul 5;10:1173616. doi: 10.3389/fmed.2023.1173616. eCollection 2023. Front Med (Lausanne). 2023. PMID: 37476610 Free PMC article.
-
Data-driven color augmentation for H&E stained images in computational pathology.J Pathol Inform. 2023 Jan 3;14:100183. doi: 10.1016/j.jpi.2022.100183. eCollection 2023. J Pathol Inform. 2023. PMID: 36687531 Free PMC article.
-
Histopathology images predict multi-omics aberrations and prognoses in colorectal cancer patients.Nat Commun. 2023 Apr 13;14(1):2102. doi: 10.1038/s41467-023-37179-4. Nat Commun. 2023. PMID: 37055393 Free PMC article.
References
LinkOut - more resources
Full Text Sources

