Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping
- PMID: 31508500
- PMCID: PMC6722120
- DOI: 10.1038/s42003-019-0570-8
Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping
Abstract
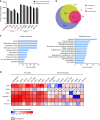
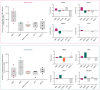
Extracellular vesicles (EVs) are a potential source of disease-associated biomarkers for diagnosis. In breast cancer, comprehensive analyses of EVs could yield robust and reliable subtype-specific biomarkers that are still critically needed to improve diagnostic routines and clinical outcome. Here, we show that proteome profiles of EVs secreted by different breast cancer cell lines are highly indicative of their respective molecular subtypes, even more so than the proteome changes within the cancer cells. Moreover, we detected molecular evidence for subtype-specific biological processes and molecular pathways, hyperphosphorylated receptors and kinases in connection with the disease, and compiled a set of protein signatures that closely reflect the associated clinical pathophysiology. These unique features revealed in our work, replicated in clinical material, collectively demonstrate the potential of secreted EVs to differentiate between breast cancer subtypes and show the prospect of their use as non-invasive liquid biopsies for diagnosis and management of breast cancer patients.
Keywords: Breast cancer; Mass spectrometry; Proteomic analysis.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures




Similar articles
-
Phosphoproteins in extracellular vesicles as candidate markers for breast cancer.Proc Natl Acad Sci U S A. 2017 Mar 21;114(12):3175-3180. doi: 10.1073/pnas.1618088114. Epub 2017 Mar 7. Proc Natl Acad Sci U S A. 2017. PMID: 28270605 Free PMC article.
-
Proteomic Profiling of Serum Extracellular Vesicles Identifies Diagnostic Signatures and Therapeutic Targets in Breast Cancer.Cancer Res. 2024 Oct 1;84(19):3267-3285. doi: 10.1158/0008-5472.CAN-23-3998. Cancer Res. 2024. PMID: 38900939 Free PMC article.
-
Extracellular vesicles from young women's breast cancer patients drive increased invasion of non-malignant cells via the Focal Adhesion Kinase pathway: a proteomic approach.Breast Cancer Res. 2020 Nov 23;22(1):128. doi: 10.1186/s13058-020-01363-x. Breast Cancer Res. 2020. PMID: 33225939 Free PMC article.
-
Protein biomarkers in breast cancer-derived extracellular vesicles for use in liquid biopsies.Am J Physiol Cell Physiol. 2021 Nov 1;321(5):C779-C797. doi: 10.1152/ajpcell.00048.2021. Epub 2021 Sep 8. Am J Physiol Cell Physiol. 2021. PMID: 34495763 Review.
-
Extracellular vesicle proteins as breast cancer biomarkers: Mass spectrometry-based analysis.Proteomics. 2024 Jun;24(11):e2300062. doi: 10.1002/pmic.202300062. Proteomics. 2024. PMID: 38829178 Review.
Cited by
-
Extracellular Vesicles Regulate Cancer Metastasis.Subcell Biochem. 2021;97:275-296. doi: 10.1007/978-3-030-67171-6_11. Subcell Biochem. 2021. PMID: 33779921
-
The proteomic analysis of breast cell line exosomes reveals disease patterns and potential biomarkers.Sci Rep. 2020 Aug 11;10(1):13572. doi: 10.1038/s41598-020-70393-4. Sci Rep. 2020. PMID: 32782317 Free PMC article.
-
Microvesicles in Cancer: Small Size, Large Potential.Int J Mol Sci. 2020 Jul 28;21(15):5373. doi: 10.3390/ijms21155373. Int J Mol Sci. 2020. PMID: 32731639 Free PMC article. Review.
-
Monitoring drug metabolic pathways through extracellular vesicles in mouse plasma.PNAS Nexus. 2024 Jan 23;3(2):pgae023. doi: 10.1093/pnasnexus/pgae023. eCollection 2024 Feb. PNAS Nexus. 2024. PMID: 38312223 Free PMC article.
-
Breast Cancer Derived Extracellular Vesicles in Bone Metastasis Induction and Their Clinical Implications as Biomarkers.Int J Mol Sci. 2020 May 18;21(10):3573. doi: 10.3390/ijms21103573. Int J Mol Sci. 2020. PMID: 32443642 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

