pathCHEMO, a generalizable computational framework uncovers molecular pathways of chemoresistance in lung adenocarcinoma
- PMID: 31508508
- PMCID: PMC6731276
- DOI: 10.1038/s42003-019-0572-6
pathCHEMO, a generalizable computational framework uncovers molecular pathways of chemoresistance in lung adenocarcinoma
Abstract
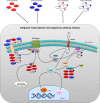
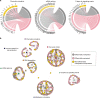
Despite recent advances in discovering a wide array of novel chemotherapy agents, identification of patients with poor and favorable chemotherapy response prior to treatment administration remains a major challenge in clinical oncology. To tackle this challenge, we present a generalizable genome-wide computational framework pathCHEMO that uncovers interplay between transcriptomic and epigenomic mechanisms altered in biological pathways that govern chemotherapy response in cancer patients. Our approach is tested on patients with lung adenocarcinoma who received adjuvant standard-of-care doublet chemotherapy (i.e., carboplatin-paclitaxel), identifying seven molecular pathway markers of primary treatment response and demonstrating their ability to predict patients at risk of carboplatin-paclitaxel resistance in an independent patient cohort (log-rank p-value = 0.008, HR = 10). Furthermore, we extend our method to additional chemotherapy-regimens and cancer types to demonstrate its accuracy and generalizability. We propose that our model can be utilized to prioritize patients for specific chemotherapy-regimens as a part of treatment planning.
Keywords: Computational models; Lung cancer; Predictive medicine.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures




Similar articles
-
A Cell-Based Method for Identification of Chemotherapy Resistance Cancer Genes.Methods Mol Biol. 2019;1907:83-90. doi: 10.1007/978-1-4939-8967-6_6. Methods Mol Biol. 2019. PMID: 30542992
-
Upregulation of Neural Precursor Cell Expressed Developmentally Downregulated 4-1 is Associated with Poor Prognosis and Chemoresistance in Lung Adenocarcinoma.Chin Med J (Engl). 2018 Jan 5;131(1):16-24. doi: 10.4103/0366-6999.221262. Chin Med J (Engl). 2018. PMID: 29271375 Free PMC article.
-
HOXB13 networking with ABCG1/EZH2/Slug mediates metastasis and confers resistance to cisplatin in lung adenocarcinoma patients.Theranostics. 2019 Apr 6;9(7):2084-2099. doi: 10.7150/thno.29463. eCollection 2019. Theranostics. 2019. PMID: 31037158 Free PMC article.
-
Predicting response to paclitaxel/carboplatin-based therapy in non-small cell lung cancer.Semin Oncol. 2001 Aug;28(4 Suppl 14):37-44. doi: 10.1016/s0093-7754(01)90058-2. Semin Oncol. 2001. PMID: 11605182 Review.
-
Overview of resistance to systemic therapy in patients with breast cancer.Adv Exp Med Biol. 2007;608:1-22. doi: 10.1007/978-0-387-74039-3_1. Adv Exp Med Biol. 2007. PMID: 17993229 Review.
Cited by
-
Big Data to Knowledge: Application of Machine Learning to Predictive Modeling of Therapeutic Response in Cancer.Curr Genomics. 2021 Dec 16;22(4):244-266. doi: 10.2174/1389202921999201224110101. Curr Genomics. 2021. PMID: 35273457 Free PMC article. Review.
-
Dynamical network analysis reveals key microRNAs in progressive stages of lung cancer.PLoS Comput Biol. 2020 May 19;16(5):e1007793. doi: 10.1371/journal.pcbi.1007793. eCollection 2020 May. PLoS Comput Biol. 2020. PMID: 32428028 Free PMC article.
-
Genome-wide analysis of therapeutic response uncovers molecular pathways governing tamoxifen resistance in ER+ breast cancer.EBioMedicine. 2020 Nov;61:103047. doi: 10.1016/j.ebiom.2020.103047. Epub 2020 Oct 21. EBioMedicine. 2020. PMID: 33099086 Free PMC article.
-
Mechanism-centric regulatory network identifies NME2 and MYC programs as markers of Enzalutamide resistance in CRPC.Nat Commun. 2024 Jan 8;15(1):352. doi: 10.1038/s41467-024-44686-5. Nat Commun. 2024. PMID: 38191557 Free PMC article.
-
Mechanism-Centric Approaches for Biomarker Detection and Precision Therapeutics in Cancer.Front Genet. 2021 Aug 2;12:687813. doi: 10.3389/fgene.2021.687813. eCollection 2021. Front Genet. 2021. PMID: 34408770 Free PMC article. Review.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA: a cancer J. Clin. 2017;67:7–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

