Benchmarking database systems for Genomic Selection implementation
- PMID: 31508797
- PMCID: PMC6737464
- DOI: 10.1093/database/baz096
Benchmarking database systems for Genomic Selection implementation
Abstract
Motivation: With high-throughput genotyping systems now available, it has become feasible to fully integrate genotyping information into breeding programs. To make use of this information effectively requires DNA extraction facilities and marker production facilities that can efficiently deploy the desired set of markers across samples with a rapid turnaround time that allows for selection before crosses needed to be made. In reality, breeders often have a short window of time to make decisions by the time they are able to collect all their phenotyping data and receive corresponding genotyping data. This presents a challenge to organize information and utilize it in downstream analyses to support decisions made by breeders. In order to implement genomic selection routinely as part of breeding programs, one would need an efficient genotyping data storage system. We selected and benchmarked six popular open-source data storage systems, including relational database management and columnar storage systems.
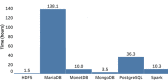
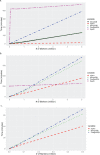
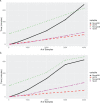
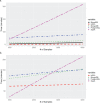
Results: We found that data extract times are greatly influenced by the orientation in which genotype data is stored in a system. HDF5 consistently performed best, in part because it can more efficiently work with both orientations of the allele matrix.
Availability: http://gobiin1.bti.cornell.edu:6083/projects/GBM/repos/benchmarking/browse.
© The Author(s) 2019. Published by Oxford University Press.
Figures

References
-
- Hickey J.M., Chiurugwi T., Mackay I. et al. (2017) Genomic prediction unifies animal and plant breeding programs to form platforms for biological discovery. Nat. Genet., 49, 1297. - PubMed
-
- Lin Z., Hayes B.J. and Daetwyler H.D. (2014) Genomic selection in crops, trees and forages: a review, Crop Pasture Sci., 65, 1177–1191.
-
- Röhm U. and Blakeley J. (2009) Data management for high-throughput genomics, arXiv Prepr. arXiv0909.1764.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

