Molecular mechanism for the multiple sclerosis risk variant rs17594362
- PMID: 31509193
- PMCID: PMC6927461
- DOI: 10.1093/hmg/ddz216
Molecular mechanism for the multiple sclerosis risk variant rs17594362
Abstract
Multiple sclerosis (MS) is known as an autoimmune demyelinating disease of the central nervous system. However, its cause remains elusive. Given previous studies suggesting that dysfunctional oligodendrocytes (OLs) may trigger MS, we tested whether single nucleotide polymorphisms (SNPs) associated with MS affect OL enhancers, potentially increasing MS risk by dysregulating gene expression of OL lineage cells. We found that two closely spaced OL enhancers, which are 3 Kb apart on chromosome 13, overlap two MS SNPs in linkage disequilibrium-rs17594362 and rs12429256. Our data revealed that the two MS SNPs significantly up-regulate the associated OL enhancers, which we have named as Rgcc-E1 and Rgcc-E2. Analysis of Hi-C data and epigenome editing experiments shows that Rgcc is the primary target of Rgcc-E1 and Rgcc-E2. Collectively, these data indicate that the molecular mechanism of rs17594362 and rs12429256 is to induce Rgcc overexpression by potentiating the enhancer activity of Rgcc-E1 and Rgcc-E2. Importantly, the dosage of the rs17594362/rs12429256 risk allele is positively correlated with the expression level of Rgcc in the human population, confirming our molecular mechanism. Our study also suggests that Rgcc overexpression in OL lineage cells may be a key cellular mechanism of rs17594362 and rs12429256 for MS.
© The Author(s) 2019. Published by Oxford University Press.
Figures
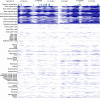

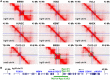

Similar articles
-
Association to the Glypican-5 gene in multiple sclerosis.J Neuroimmunol. 2010 Sep 14;226(1-2):194-7. doi: 10.1016/j.jneuroim.2010.07.003. Epub 2010 Aug 6. J Neuroimmunol. 2010. PMID: 20692050
-
Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits.Genome Res. 2014 Jan;24(1):1-13. doi: 10.1101/gr.164079.113. Epub 2013 Nov 6. Genome Res. 2014. PMID: 24196873 Free PMC article.
-
Multiple Functional Variants at 13q14 Risk Locus for Osteoporosis Regulate RANKL Expression Through Long-Range Super-Enhancer.J Bone Miner Res. 2018 Jul;33(7):1335-1346. doi: 10.1002/jbmr.3419. Epub 2018 May 17. J Bone Miner Res. 2018. PMID: 29528523
-
Susceptibility variants in the CD58 gene locus point to a role of microRNA-548ac in the pathogenesis of multiple sclerosis.Mutat Res Rev Mutat Res. 2015 Jan-Mar;763:161-7. doi: 10.1016/j.mrrev.2014.10.002. Epub 2014 Oct 16. Mutat Res Rev Mutat Res. 2015. PMID: 25795118 Review.
-
From genetic associations to functional studies in multiple sclerosis.Eur J Neurol. 2016 May;23(5):847-53. doi: 10.1111/ene.12981. Epub 2016 Mar 7. Eur J Neurol. 2016. PMID: 26948534 Review.
Cited by
-
A systematic strategy for identifying causal single nucleotide polymorphisms and their target genes on Juvenile arthritis risk haplotypes.BMC Med Genomics. 2024 Jul 12;17(1):185. doi: 10.1186/s12920-024-01954-z. BMC Med Genomics. 2024. PMID: 38997781 Free PMC article.
-
Identifying an oligodendrocyte enhancer that regulates Olig2 expression.Hum Mol Genet. 2023 Feb 19;32(5):835-846. doi: 10.1093/hmg/ddac249. Hum Mol Genet. 2023. PMID: 36193754 Free PMC article.
-
Identifying oligodendrocyte enhancers governing Plp1 expression.Hum Mol Genet. 2021 Nov 16;30(23):2225-2239. doi: 10.1093/hmg/ddab184. Hum Mol Genet. 2021. PMID: 34230963 Free PMC article.
-
RGCC balances self-renewal and neuronal differentiation of neural stem cells in the developing mammalian neocortex.EMBO Rep. 2021 Sep 6;22(9):e51781. doi: 10.15252/embr.202051781. Epub 2021 Jul 29. EMBO Rep. 2021. PMID: 34323349 Free PMC article.
-
Uncovering oligodendrocyte enhancers that control Cnp expression.Hum Mol Genet. 2023 Nov 17;32(23):3225-3236. doi: 10.1093/hmg/ddad141. Hum Mol Genet. 2023. PMID: 37642363 Free PMC article.
References
-
- Trapp B.D. and Nave K.-A. (2008) Multiple sclerosis: an immune or neurodegenerative disorder? Annu. Rev. Neurosci., 31, 247–269. - PubMed
-
- Hauser S.L., Chan J.R. and Oksenberg J.R. (2013) Multiple sclerosis: prospects and promise. Ann. Neurol., 74, 317–327. - PubMed
-
- Dendrou C.A., Fugger L. and Friese M.A. (2015) Immunopathology of multiple sclerosis. Nat. Rev. Immunol., 15, 545–558. - PubMed
-
- Nakahara J., Maeda M., Aiso S. and Suzuki N. (2012) Current concepts in multiple sclerosis: autoimmunity versus oligodendrogliopathy. Clin. Rev. Allergy Immunol., 42, 26–34. - PubMed
-
- Stys P.K., Zamponi G.W., van Minnen J. and Geurts J.J.G. (2012) Will the real multiple sclerosis please stand up? Nat. Rev. Neurosci., 13, 507–514. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

