iTRAQ-based proteomic analysis reveals key proteins affecting cardiac function in broilers that died of sudden death syndrome
- PMID: 31509194
- PMCID: PMC8913949
- DOI: 10.3382/ps/pez532
iTRAQ-based proteomic analysis reveals key proteins affecting cardiac function in broilers that died of sudden death syndrome
Abstract
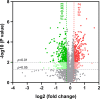
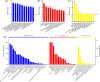
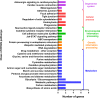
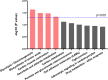
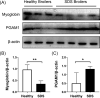
Sudden death syndrome (SDS), which is a cardiac-related condition commonly observed in chickens selected for rapid growth, causes significant economic losses to the global poultry industry. Its pathogenesis in broilers is poorly understood, and little is known about the proteome of the heart tissue of SDS broilers. A quantitative proteomic approach using isobaric tags for relative and absolute quantification labeling of peptides was used to characterize the protein expression profiles in the left ventricle of SDS broilers. These proteins were further analyzed by bioinformatics, and two proteins were validated by western blot analysis. We identified 186 differentially expressed proteins (DEPs), of which 72 were upregulated, and 114 were downregulated in the SDS group. Functional annotation suggested that 7 DEPs were related to cardiac muscle contraction, and another 7 DEPs were related to cardiac energy metabolism. Protein interaction network predictions indicated that differences in cardiac muscle contraction between SDS and healthy groups were regulated by troponin T, tropomyosin alpha-1 chain, fast myosin heavy chain HCIII, myosin-1B, coronin, and myoglobin, whereas differences in cardiac energy metabolism and biosynthesis of amino acids were regulated by gamma-enolase, phosphoglycerate mutase, NADH-ubiquinone oxidoreductase chain 2, serine/threonine-protein kinase, myoglobin, and alpha-amylase. Our expression profiles provide useful information and new insights into key proteins to elucidate SDS for further studies.
Keywords: broiler chicken; gene ontology analysis; iTRAQ analysis; sudden death syndrome.
© 2019 Poultry Science Association Inc.
Figures
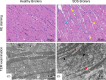
 ) indicates the inflammatory cells. Yellow arrows (
) indicates the inflammatory cells. Yellow arrows ( ) indicate the myocardial cells showed obvious granular degeneration. (C and D) Electron microscopy pictures showing the differences in myocardial fibers and mitochondria between healthy broilers and SDS broilers (×10,000). Red arrows (
) indicate the myocardial cells showed obvious granular degeneration. (C and D) Electron microscopy pictures showing the differences in myocardial fibers and mitochondria between healthy broilers and SDS broilers (×10,000). Red arrows ( ) indicate the myocardial fibers of SDS broilers were broken, edema and irregular arrangement. Black arrows (→) indicate the damaged mitochondria.
) indicate the myocardial fibers of SDS broilers were broken, edema and irregular arrangement. Black arrows (→) indicate the damaged mitochondria.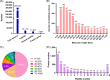

Similar articles
-
A study on pathogenesis of sudden death syndrome in broiler chickens.Res Vet Sci. 2008 Aug;85(1):131-40. doi: 10.1016/j.rvsc.2007.08.006. Epub 2007 Sep 27. Res Vet Sci. 2008. PMID: 17904171
-
Role of cardiac hypoxia in the pathogenesis of sudden death syndrome in broiler chickens - A metabolic and molecular study.Acta Vet Hung. 2021 Mar 19;69(1):43-49. doi: 10.1556/004.2021.00004. Acta Vet Hung. 2021. PMID: 33764895
-
Polymorphism identification and cardiac gene expression analysis of the calsequestrin 2 gene in broiler chickens with sudden death syndrome.Br Poult Sci. 2016 Apr;57(2):151-60. doi: 10.1080/00071668.2015.1099615. Br Poult Sci. 2016. PMID: 26953612
-
Sequence and expression analysis of cardiac ryanodine receptor 2 in broilers that died from sudden death syndrome.Avian Pathol. 2019 Oct;48(5):444-453. doi: 10.1080/03079457.2019.1618439. Epub 2019 Jun 25. Avian Pathol. 2019. PMID: 31081346
-
A consideration of comparative metabolic aspects of the aetiology of sudden death syndrome and ascites in broilers.Br Vet J. 1993 May-Jun;149(3):285-94. doi: 10.1016/s0007-1935(05)80174-5. Br Vet J. 1993. PMID: 8334510 Review.
Cited by
-
Proteomics study on the effect of silybin on cardiomyopathy in obese mice.Sci Rep. 2021 Mar 30;11(1):7136. doi: 10.1038/s41598-021-86717-x. Sci Rep. 2021. PMID: 33785854 Free PMC article.
-
Quantitative proteomic analysis of chicken serum reveals key proteins affecting follicle development during reproductive phase transitions.Poult Sci. 2021 Jan;100(1):325-333. doi: 10.1016/j.psj.2020.09.058. Epub 2020 Oct 8. Poult Sci. 2021. PMID: 33357697 Free PMC article.
References
-
- Aggarwal S., Yadav A.K. Dissecting the iTRAQ data analysis. Methods Mol. Biol. 2016;1362:277–291. - PubMed
-
- Ai C., Kong L. CGPS: a machine learning-based approach integrating multiple gene set analysis tools for better prioritization of biologically relevant pathways. J. Genet. Genomics. 2018;45:489–504. - PubMed
-
- Basaki M., Asasi K., Tabandeh M.R., Aminlari M. Polymorphism identification and cardiac gene expression analysis of the calsequestrin 2 gene in broiler chickens with sudden death syndrome. Br. Poult. Sci. 2016;57:151–160. - PubMed
-
- Bers D.M. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. - PubMed

