Shortened nuclear matrix attachment regions are sufficient for replication and maintenance of episomes in mammalian cells
- PMID: 31509492
- PMCID: PMC6789156
- DOI: 10.1091/mbc.E19-02-0108
Shortened nuclear matrix attachment regions are sufficient for replication and maintenance of episomes in mammalian cells
Abstract
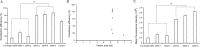
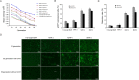
Matrix attachment regions (MARs) can mediate the replication of vector episomes in mammalian cells; however, the molecular mode of action remains unclear. Here, we assessed the characteristics of MARs and the mechanism that mediates episomal vector replication in mammalian cells. Five shortened subfragments of β-interferon MAR fragments were cloned and transferred into CHO cells, and transgene expression levels, presence of the gene, and the episomal maintenance mechanism were determined. Three shortened MAR derivatives (position 781-1320, 1201-1740, and 1621-2201) retained full MAR activity and mediated episomal vector replication. Moreover, the three shortened MARs showed higher transgene expression levels, greater efficiency in colony formation, and more persistent transgene expression compared with those of the original pEPI-1 plasmid, and three functional truncated MARs can bind to SAF-A MAR-binding protein. These results suggest that shortened MARs are sufficient for replication and maintenance of episomes in CHO cells.
Figures




Similar articles
-
MAR characteristic motifs mediate episomal vector in CHO cells.Gene. 2015 Apr 1;559(2):137-43. doi: 10.1016/j.gene.2015.01.032. Epub 2015 Jan 15. Gene. 2015. PMID: 25598284
-
The functional role of S/MARs in episomal vectors as defined by the stress-induced destabilization profile of the vector sequences.J Mol Biol. 2009 Apr 17;387(5):1239-49. doi: 10.1016/j.jmb.2009.02.043. Epub 2009 Feb 25. J Mol Biol. 2009. PMID: 19248788
-
Enhancing expression level and stability of transgene mediated by episomal vector via buffering DNA methyltransferase in transfected CHO cells.J Cell Biochem. 2019 Sep;120(9):15661-15670. doi: 10.1002/jcb.28835. Epub 2019 May 9. J Cell Biochem. 2019. PMID: 31074065
-
Sustained transgene expression using MAR elements.Curr Gene Ther. 2008 Oct;8(5):353-66. doi: 10.2174/156652308786071032. Curr Gene Ther. 2008. PMID: 18855632 Review.
-
Non-viral episomal modification of cells using S/MAR elements.Expert Opin Biol Ther. 2011 Sep;11(9):1177-91. doi: 10.1517/14712598.2011.582035. Epub 2011 May 9. Expert Opin Biol Ther. 2011. PMID: 21548848 Review.
Cited by
-
Revisiting the unique structure of autonomously replicating sequences in Yarrowia lipolytica and its role in pathway engineering.Appl Microbiol Biotechnol. 2021 Aug;105(14-15):5959-5972. doi: 10.1007/s00253-021-11399-4. Epub 2021 Aug 6. Appl Microbiol Biotechnol. 2021. PMID: 34357429
-
A three-stage assembly program governing pancreatic, plasma, pituitary, and bone secretory cell differentiation: A strategy to augment protein delivery.J Biol Chem. 2025 Aug 5;301(9):110562. doi: 10.1016/j.jbc.2025.110562. Online ahead of print. J Biol Chem. 2025. PMID: 40774385 Free PMC article. Review.
-
Improved Functionality of Integration-Deficient Lentiviral Vectors (IDLVs) by the Inclusion of IS2 Protein Docks.Pharmaceutics. 2021 Aug 6;13(8):1217. doi: 10.3390/pharmaceutics13081217. Pharmaceutics. 2021. PMID: 34452178 Free PMC article.
-
Advances in the Development and the Applications of Nonviral, Episomal Vectors for Gene Therapy.Hum Gene Ther. 2021 Oct;32(19-20):1076-1095. doi: 10.1089/hum.2020.310. Epub 2021 Sep 20. Hum Gene Ther. 2021. PMID: 34348480 Free PMC article.
-
Minicircle Delivery to the Neural Retina as a Gene Therapy Approach.Int J Mol Sci. 2022 Oct 2;23(19):11673. doi: 10.3390/ijms231911673. Int J Mol Sci. 2022. PMID: 36232975 Free PMC article. Review.
References
-
- Argyros O, Wong SP, Fedonidis C, Tolmachov O, Waddington SN, Howe SJ, Niceta M, Coutelle C, Harbottle RP. (2011a). Development of S/MAR minicircles for enhanced and persistent transgene expression in the mouse liver. J Mol Med (Berl) , 515–529. - PubMed
-
- Argyros O, Wong SP, Harbottle RP. (2011b). Non-viral episomal modification of cells using S/MAR elements. Expert Opin Biol Ther , 1177–1191. - PubMed
-
- Baiker A, Maercker C, Piechaczek C, Schmidt SB, Bode J, Benham C, Lipps HJ. (2000). Mitotic stability of an episomal vector containing a human scaffold/matrix-attached region is provided by association with nuclear matrix. Nat Cell Biol , 182–184. - PubMed
-
- Broll S, Oumard A, Hahn K, Schambach A, Bode J. (2010). Minicircle performance depending on S/MAR-nuclear matrix interactions. J Mol Biol , 950–965. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

