Demonstrating specificity of bioactive peptide nucleic acids (PNAs) targeting microRNAs for practical laboratory classes of applied biochemistry and pharmacology
- PMID: 31509554
- PMCID: PMC6738603
- DOI: 10.1371/journal.pone.0221923
Demonstrating specificity of bioactive peptide nucleic acids (PNAs) targeting microRNAs for practical laboratory classes of applied biochemistry and pharmacology
Abstract
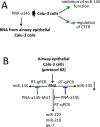
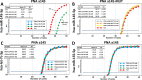
Practical laboratory classes teaching molecular pharmacology approaches employed in the development of therapeutic strategies are of great interest for students of courses in Biotechnology, Applied Biology, Pharmaceutic and Technology Chemistry, Translational Oncology. Unfortunately, in most cases the technology to be transferred to learning students is complex and requires multi-step approaches. In this respect, simple and straightforward experimental protocols might be of great interest. This study was aimed at presenting a laboratory exercise focusing (a) on a very challenging therapeutic strategy, i.e. microRNA therapeutics, and (b) on the employment of biomolecules of great interest in applied biology and pharmacology, i.e. peptide nucleic acids (PNAs). The aims of the practical laboratory were to determine: (a) the possible PNA-mediated arrest in RT-qPCR, to be eventually used to demonstrate PNA targeting of selected miRNAs; (b) the possible lack of activity on mutated PNA sequences; (c) the effects (if any) on the amplification of other unrelated miRNA sequences. The results which can be obtained support the following conclusions: PNA-mediated arrest in RT-qPCR can be analyzed in a easy way; mutated PNA sequences are completely inactive; the effects of the employed PNAs are specific and no inhibitory effect occurs on other unrelated miRNA sequences. This activity is simple (cell culture, RNA extraction, RT-qPCR are all well-established technologies), fast (starting from isolated and characterized RNA, few hours are just necessary), highly reproducible (therefore easily employed by even untrained students). On the other hand, these laboratory lessons require some facilities, the most critical being the availability of instruments for PCR. While this might be a problem in the case these instruments are not available, we would like to underline that determination of the presence or of a lack of amplified product can be also obtained using standard analytical approaches based on agarose gel electrophoresis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Teaching during COVID-19 pandemic in practical laboratory classes of applied biochemistry and pharmacology: A validated fast and simple protocol for detection of SARS-CoV-2 Spike sequences.PLoS One. 2022 Apr 6;17(4):e0266419. doi: 10.1371/journal.pone.0266419. eCollection 2022. PLoS One. 2022. PMID: 35385518 Free PMC article.
-
A Peptide Nucleic Acid against MicroRNA miR-145-5p Enhances the Expression of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) in Calu-3 Cells.Molecules. 2017 Dec 29;23(1):71. doi: 10.3390/molecules23010071. Molecules. 2017. PMID: 29286300 Free PMC article.
-
A highly effective and long-lasting inhibition of miRNAs with PNA-based antisense oligonucleotides.Mol Cells. 2009 Oct 31;28(4):341-5. doi: 10.1007/s10059-009-0134-8. Epub 2009 Sep 30. Mol Cells. 2009. PMID: 19812898
-
Peptide-nucleic acids (PNAs): a tool for the development of gene expression modifiers.Curr Pharm Des. 2001 Nov;7(17):1839-62. doi: 10.2174/1381612013397087. Curr Pharm Des. 2001. PMID: 11562312 Review.
-
miRNA therapeutics: delivery and biological activity of peptide nucleic acids targeting miRNAs.Epigenomics. 2011 Dec;3(6):733-45. doi: 10.2217/epi.11.90. Epigenomics. 2011. PMID: 22126292 Review.
Cited by
-
MicroRNAs miR-584-5p and miR-425-3p Are Up-Regulated in Plasma of Colorectal Cancer (CRC) Patients: Targeting with Inhibitor Peptide Nucleic Acids Is Associated with Induction of Apoptosis in Colon Cancer Cell Lines.Cancers (Basel). 2022 Dec 25;15(1):128. doi: 10.3390/cancers15010128. Cancers (Basel). 2022. PMID: 36612125 Free PMC article.
-
Teaching during COVID-19 pandemic in practical laboratory classes of applied biochemistry and pharmacology: A validated fast and simple protocol for detection of SARS-CoV-2 Spike sequences.PLoS One. 2022 Apr 6;17(4):e0266419. doi: 10.1371/journal.pone.0266419. eCollection 2022. PLoS One. 2022. PMID: 35385518 Free PMC article.
-
A Peptide Nucleic Acid (PNA) Masking the miR-145-5p Binding Site of the 3'UTR of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) mRNA Enhances CFTR Expression in Calu-3 Cells.Molecules. 2020 Apr 5;25(7):1677. doi: 10.3390/molecules25071677. Molecules. 2020. PMID: 32260566 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

