Histone Methylation Participates in Gene Expression Control during the Early Development of the Pacific Oyster Crassostrea gigas
- PMID: 31509985
- PMCID: PMC6771004
- DOI: 10.3390/genes10090695
Histone Methylation Participates in Gene Expression Control during the Early Development of the Pacific Oyster Crassostrea gigas
Abstract
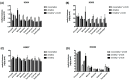
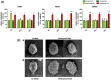
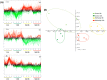
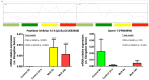
Histone methylation patterns are important epigenetic regulators of mammalian development, notably through stem cell identity maintenance by chromatin remodeling and transcriptional control of pluripotency genes. But, the implications of histone marks are poorly understood in distant groups outside vertebrates and ecdysozoan models. However, the development of the Pacific oyster Crassostrea gigas is under the strong epigenetic influence of DNA methylation, and Jumonji histone-demethylase orthologues are highly expressed during C. gigas early life. This suggests a physiological relevance of histone methylation regulation in oyster development, raising the question of functional conservation of this epigenetic pathway in lophotrochozoan. Quantification of histone methylation using fluorescent ELISAs during oyster early life indicated significant variations in monomethyl histone H3 lysine 4 (H3K4me), an overall decrease in H3K9 mono- and tri-methylations, and in H3K36 methylations, respectively, whereas no significant modification could be detected in H3K27 methylation. Early in vivo treatment with the JmjC-specific inhibitor Methylstat induced hypermethylation of all the examined histone H3 lysines and developmental alterations as revealed by scanning electronic microscopy. Using microarrays, we identified 376 genes that were differentially expressed under methylstat treatment, which expression patterns could discriminate between samples as indicated by principal component analysis. Furthermore, Gene Ontology revealed that these genes were related to processes potentially important for embryonic stages such as binding, cell differentiation and development. These results suggest an important physiological significance of histone methylation in the oyster embryonic and larval life, providing, to our knowledge, the first insights into epigenetic regulation by histone methylation in lophotrochozoan development.
Keywords: H3K27; H3K36; H3K4; H3K9; embryos; epigenetics; histone modifications; methylstat; mollusk.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
Temperature influences histone methylation and mRNA expression of the Jmj-C histone-demethylase orthologues during the early development of the oyster Crassostrea gigas.Mar Genomics. 2015 Feb;19:23-30. doi: 10.1016/j.margen.2014.09.002. Epub 2014 Sep 16. Mar Genomics. 2015. PMID: 25224965
-
The Jumonji gene family in Crassostrea gigas suggests evolutionary conservation of Jmj-C histone demethylases orthologues in the oyster gametogenesis and development.Gene. 2014 Mar 15;538(1):164-75. doi: 10.1016/j.gene.2013.12.016. Epub 2014 Jan 6. Gene. 2014. PMID: 24406622
-
A functional m6 A-RNA methylation pathway in the oyster Crassostrea gigas assumes epitranscriptomic regulation of lophotrochozoan development.FEBS J. 2021 Mar;288(5):1696-1711. doi: 10.1111/febs.15500. Epub 2020 Sep 1. FEBS J. 2021. PMID: 32743927
-
Epigenetic regulation: methylation of histone and non-histone proteins.Sci China C Life Sci. 2009 Apr;52(4):311-22. doi: 10.1007/s11427-009-0054-z. Epub 2009 Apr 21. Sci China C Life Sci. 2009. PMID: 19381457 Review.
-
Histone modification and the control of heterochromatic gene silencing in Drosophila.Chromosome Res. 2006;14(4):377-92. doi: 10.1007/s10577-006-1066-1. Chromosome Res. 2006. PMID: 16821134 Review.
Cited by
-
Updates on the role of epigenetics in familial mediterranean fever (FMF).Orphanet J Rare Dis. 2024 Feb 26;19(1):90. doi: 10.1186/s13023-024-03098-w. Orphanet J Rare Dis. 2024. PMID: 38409042 Free PMC article. Review.
-
Epigenetic machinery is functionally conserved in cephalopods.BMC Biol. 2022 Sep 14;20(1):202. doi: 10.1186/s12915-022-01404-1. BMC Biol. 2022. PMID: 36104784 Free PMC article.
-
Concomitant downregulation of neuropeptide genes in a marine snail with consecutive sexual maturation after a nuclear disaster in Japan.Front Endocrinol (Lausanne). 2023 Mar 10;14:1129666. doi: 10.3389/fendo.2023.1129666. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 36967776 Free PMC article.
-
Core histone families of mollusca: systematic identification, evolutionary insights, and functional analysis.BMC Genomics. 2025 Jul 1;26(1):594. doi: 10.1186/s12864-025-11776-7. BMC Genomics. 2025. PMID: 40596809 Free PMC article.
-
The Modification of H3K4me3 Enhanced the Expression of CgTLR3 in Hemocytes to Increase CgIL17-1 Production in the Immune Priming of Crassostrea gigas.Int J Mol Sci. 2024 Jan 15;25(2):1036. doi: 10.3390/ijms25021036. Int J Mol Sci. 2024. PMID: 38256110 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

