MicroRNA Biogenesis Pathway Genes Are Deregulated in Colorectal Cancer
- PMID: 31510013
- PMCID: PMC6770105
- DOI: 10.3390/ijms20184460
MicroRNA Biogenesis Pathway Genes Are Deregulated in Colorectal Cancer
Abstract
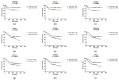
MicroRNAs (miRNAs) are small non-coding RNAs that post-transcriptionally regulate gene expression. Each step of their production and maturation has to be strictly regulated, as any disruption of control mechanisms may lead to cancer. Thus, we have measured the expression of 19 genes involved in miRNAs biogenesis pathway in tumor tissues of 239 colorectal cancer (CRC) patients, 17 CRC patients with liver metastases and 239 adjacent tissues using real-time PCR. Subsequently, the expression of analyzed genes was correlated with the clinical-pathological features as well as with the survival of patients. In total, significant over-expression of all analyzed genes was observed in tumor tissues as well as in liver metastases except for LIN28A/B. Furthermore, it was shown that the deregulated levels of some of the analyzed genes significantly correlate with tumor stage, grade, location, size and lymph node positivity. Finally, high levels of DROSHA and TARBP2 were associated with shorter disease-free survival, while the over-expression of XPO5, TNRC6A and DDX17 was detected in tissues of patients with shorter overall survival and poor prognosis. Our data indicate that changed levels of miRNA biogenesis genes may contribute to origin as well as progression of CRC; thus, these molecules could serve as potential therapeutic targets.
Keywords: RT-qPCR; biogenesis; colorectal cancer; disease-free survival; microRNA; overall survival.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

