Effects of Microbial Activity and Environmental Parameters on the Degradation of Extracellular Environmental DNA from a Eutrophic Lake
- PMID: 31510040
- PMCID: PMC6765872
- DOI: 10.3390/ijerph16183339
Effects of Microbial Activity and Environmental Parameters on the Degradation of Extracellular Environmental DNA from a Eutrophic Lake
Abstract
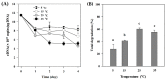
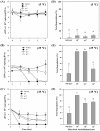
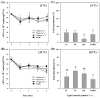
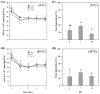
Extracellular DNA (exDNA) pool in aquatic environments is a valuable source for biomonitoring and bioassessment. However, degradation under particular environmental conditions can hamper exDNA detectability over time. In this study, we analyzed how different biotic and abiotic factors affect the degradation rate of extracellular environmental DNA using 16S rDNA sequences extracted from the sediment of a eutrophic lake and Anabaena variabilis cultured in the laboratory. We exposed the extracted exDNA to different levels of temperature, light, pH, and bacterial activity, and quantitatively analyzed the concentration of exDNA during 4 days. The solution containing bacteria for microbial activity treatment was obtained from the lake sediment using four consecutive steps of filtration; two mesh filters (100 μm and 60 μm mesh) and two glass fiber filters (2.7 μm and 1.2 μm pore-sized). We found that temperature individually and in combination with bacterial abundance had significant positive effects on the degradation of exDNA. The highest degradation rate was observed in samples exposed to high microbial activity, where exDNA was completely degraded within 1 day at a rate of 3.27 day-1. Light intensity and pH had no significant effects on degradation rate of exDNA. Our results indicate that degradation of exDNA in freshwater ecosystems is driven by the combination of both biotic and abiotic factors and it may occur very fast under particular conditions.
Keywords: biomonitoring; degradation; extracellular DNA; freshwater environment; light; microbial activity; pH; temperature.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Preservation of microbial DNA in marine sediments: insights from extracellular DNA pools.Environ Microbiol. 2018 Dec;20(12):4526-4542. doi: 10.1111/1462-2920.14401. Epub 2018 Oct 5. Environ Microbiol. 2018. PMID: 30198168
-
Ammonia-oxidizing archaea and bacteria in water columns and sediments of a highly eutrophic plateau freshwater lake.Environ Sci Pollut Res Int. 2016 Aug;23(15):15358-69. doi: 10.1007/s11356-016-6707-0. Epub 2016 Apr 25. Environ Sci Pollut Res Int. 2016. PMID: 27109114
-
Abundance and diversity of iron reducing bacteria communities in the sediments of a heavily polluted freshwater lake.Appl Microbiol Biotechnol. 2018 Dec;102(24):10791-10801. doi: 10.1007/s00253-018-9443-1. Epub 2018 Oct 17. Appl Microbiol Biotechnol. 2018. PMID: 30334090
-
Extracellular DNA in natural environments: features, relevance and applications.Appl Microbiol Biotechnol. 2018 Aug;102(15):6343-6356. doi: 10.1007/s00253-018-9120-4. Epub 2018 Jun 1. Appl Microbiol Biotechnol. 2018. PMID: 29858957 Free PMC article. Review.
-
Rapid progression and future of environmental DNA research.Commun Biol. 2019 Feb 27;2:80. doi: 10.1038/s42003-019-0330-9. eCollection 2019. Commun Biol. 2019. PMID: 30820475 Free PMC article. Review.
Cited by
-
The Use of Environmental DNA as Preliminary Description of Invertebrate Diversity in Three Sicilian Lakes.Animals (Basel). 2025 Jan 26;15(3):355. doi: 10.3390/ani15030355. Animals (Basel). 2025. PMID: 39943125 Free PMC article.
-
Eutrophication and the Ecological Health Risk.Int J Environ Res Public Health. 2020 Aug 31;17(17):6332. doi: 10.3390/ijerph17176332. Int J Environ Res Public Health. 2020. PMID: 32878106 Free PMC article.
-
The Multiple States of Environmental DNA and What Is Known about Their Persistence in Aquatic Environments.Environ Sci Technol. 2022 May 3;56(9):5322-5333. doi: 10.1021/acs.est.1c07638. Epub 2022 Apr 18. Environ Sci Technol. 2022. PMID: 35435663 Free PMC article. Review.
-
Potential Role of Photochemistry in Environmental DNA Degradation.Environ Sci Technol Lett. 2024 Nov 26;11(12):1284-1295. doi: 10.1021/acs.estlett.4c00704. eCollection 2024 Dec 10. Environ Sci Technol Lett. 2024. PMID: 39678710 Free PMC article. Review.
-
Water pre-filtration methods to improve environmental DNA detection by real-time PCR and metabarcoding.PLoS One. 2021 May 7;16(5):e0250162. doi: 10.1371/journal.pone.0250162. eCollection 2021. PLoS One. 2021. PMID: 33961651 Free PMC article.
References
-
- Keller S.R., Hilderbrand R.H., Shank M.K., Potapova M. Environmental DNA genetic monitoring of the nuisance freshwater diatom, Didymosphenia geminata, in eastern North American streams. Divers. Distrib. 2017;23:381–393. doi: 10.1111/ddi.12536. - DOI
-
- Foote A.D., Thomsen P.F., Sveegaard S., Wahlberg M., Kielgast J., Kyhn L.A., Salling A.B., Galatius A., Orlando L., Gilbert M.T.P. Investigating the potential use of environmental DNA (eDNA) for genetic monitoring of marine mammals. PLoS ONE. 2012;7:e41781. doi: 10.1371/journal.pone.0041781. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

