Controlling large Boolean networks with single-step perturbations
- PMID: 31510648
- PMCID: PMC6612811
- DOI: 10.1093/bioinformatics/btz371
Controlling large Boolean networks with single-step perturbations
Abstract
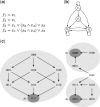
Motivation: The control of Boolean networks has traditionally focussed on strategies where the perturbations are applied to the nodes of the network for an extended period of time. In this work, we study if and how a Boolean network can be controlled by perturbing a minimal set of nodes for a single-step and letting the system evolve afterwards according to its original dynamics. More precisely, given a Boolean network (BN), we compute a minimal subset Cmin of the nodes such that BN can be driven from any initial state in an attractor to another 'desired' attractor by perturbing some or all of the nodes of Cmin for a single-step. Such kind of control is attractive for biological systems because they are less time consuming than the traditional strategies for control while also being financially more viable. However, due to the phenomenon of state-space explosion, computing such a minimal subset is computationally inefficient and an approach that deals with the entire network in one-go, does not scale well for large networks.
Results: We develop a 'divide-and-conquer' approach by decomposing the network into smaller partitions, computing the minimal control on the projection of the attractors to these partitions and then composing the results to obtain Cmin for the whole network. We implement our method and test it on various real-life biological networks to demonstrate its applicability and efficiency.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2019. Published by Oxford University Press.
Figures
References
-
- Adai A.T. et al. (2004) LGL: creating a map of protein function with an algorithm for visualizing very large biological networks. J. Mol. Biol., 340, 179–190. - PubMed
-
- Cervera L. et al. (2015) Extended gene expression by medium exchange and repeated transient transfection for recombinant protein production enhancement. Biotechnol. Bioeng., 112, 934–946. - PubMed