In vivo epigenetic editing of Sema6a promoter reverses transcallosal dysconnectivity caused by C11orf46/Arl14ep risk gene
- PMID: 31511512
- PMCID: PMC6739341
- DOI: 10.1038/s41467-019-12013-y
In vivo epigenetic editing of Sema6a promoter reverses transcallosal dysconnectivity caused by C11orf46/Arl14ep risk gene
Abstract
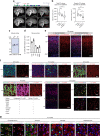
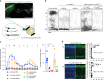
Many neuropsychiatric risk genes contribute to epigenetic regulation but little is known about specific chromatin-associated mechanisms governing the formation of neuronal connectivity. Here we show that transcallosal connectivity is critically dependent on C11orf46, a nuclear protein encoded in the chromosome 11p13 WAGR risk locus. C11orf46 haploinsufficiency was associated with hypoplasia of the corpus callosum. C11orf46 knockdown disrupted transcallosal projections and was rescued by wild type C11orf46 but not the C11orf46R236H mutant associated with intellectual disability. Multiple genes encoding key regulators of axonal development, including Sema6a, were hyperexpressed in C11orf46-knockdown neurons. RNA-guided epigenetic editing of Sema6a gene promoters via a dCas9-SunTag system with C11orf46 binding normalized SEMA6A expression and rescued transcallosal dysconnectivity via repressive chromatin remodeling by the SETDB1 repressor complex. Our study demonstrates that interhemispheric communication is sensitive to locus-specific remodeling of neuronal chromatin, revealing the therapeutic potential for shaping the brain's connectome via gene-targeted designer activators and repressor proteins.
Conflict of interest statement
J.C.H. is the recipient of an unrestricted research grant from Rhythm Pharmaceuticals. The other authors declare no competing interests.
Figures



Similar articles
-
dCas9-based epigenome editing suggests acquisition of histone methylation is not sufficient for target gene repression.Nucleic Acids Res. 2017 Sep 29;45(17):9901-9916. doi: 10.1093/nar/gkx578. Nucleic Acids Res. 2017. PMID: 28973434 Free PMC article.
-
Stabilization of Foxp3 expression by CRISPR-dCas9-based epigenome editing in mouse primary T cells.Epigenetics Chromatin. 2017 May 8;10:24. doi: 10.1186/s13072-017-0129-1. eCollection 2017. Epigenetics Chromatin. 2017. PMID: 28503202 Free PMC article.
-
Barrier-to-Autointegration Factor 1 (BAF/BANF1) Promotes Association of the SETD1A Histone Methyltransferase with Herpes Simplex Virus Immediate-Early Gene Promoters.mBio. 2015 May 26;6(3):e00345-15. doi: 10.1128/mBio.00345-15. mBio. 2015. PMID: 26015494 Free PMC article.
-
A review on CRISPR/Cas-based epigenetic regulation in plants.Int J Biol Macromol. 2022 Oct 31;219:1261-1271. doi: 10.1016/j.ijbiomac.2022.08.182. Epub 2022 Aug 31. Int J Biol Macromol. 2022. PMID: 36057300 Review.
-
Epigenome editing: targeted manipulation of epigenetic modifications in plants.Genes Genomics. 2022 Mar;44(3):307-315. doi: 10.1007/s13258-021-01199-5. Epub 2022 Jan 9. Genes Genomics. 2022. PMID: 35000141 Review.
Cited by
-
Integrative computational epigenomics to build data-driven gene regulation hypotheses.Gigascience. 2020 Jun 1;9(6):giaa064. doi: 10.1093/gigascience/giaa064. Gigascience. 2020. PMID: 32543653 Free PMC article.
-
SETDB1-like MET-2 promotes transcriptional silencing and development independently of its H3K9me-associated catalytic activity.Nat Struct Mol Biol. 2022 Feb;29(2):85-96. doi: 10.1038/s41594-021-00712-4. Epub 2022 Jan 31. Nat Struct Mol Biol. 2022. PMID: 35102319 Free PMC article.
-
Unusual Presentation in WAGR Syndrome: Expanding the Phenotypic and Genotypic Spectrum of the Diseases.Genes (Basel). 2022 Aug 12;13(8):1431. doi: 10.3390/genes13081431. Genes (Basel). 2022. PMID: 36011342 Free PMC article.
-
Roles of eIF3m in the tumorigenesis of triple negative breast cancer.Cancer Cell Int. 2020 Apr 29;20:141. doi: 10.1186/s12935-020-01220-z. eCollection 2020. Cancer Cell Int. 2020. PMID: 32368187 Free PMC article.
-
Single-base m6A epitranscriptomics reveals novel HIV-1 host interaction targets in primary CD4+ T cells.bioRxiv [Preprint]. 2025 Jan 2:2024.12.31.630958. doi: 10.1101/2024.12.31.630958. bioRxiv. 2025. PMID: 39803509 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

