Prognostic and predictive enrichment in sepsis
- PMID: 31511662
- PMCID: PMC7097452
- DOI: 10.1038/s41581-019-0199-3
Prognostic and predictive enrichment in sepsis
Abstract
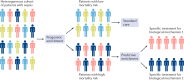
Sepsis is a heterogeneous disease state that is both common and consequential in critically ill patients. Unfortunately, the heterogeneity of sepsis at the individual patient level has hindered advances in the field beyond the current therapeutic standards, which consist of supportive care and antibiotics. This complexity has prompted attempts to develop a precision medicine approach, with research aimed towards stratifying patients into more homogeneous cohorts with shared biological features, potentially facilitating the identification of new therapies. Several investigators have successfully utilized leukocyte-derived mRNA and discovery-based approaches to subgroup patients on the basis of biological similarities defined by transcriptomic signatures. A critical next step is to develop a consensus sepsis subclassification system, which includes transcriptomic signatures as well as other biological and clinical data. This goal will require collaboration among various investigative groups, and validation in both existing data sets and prospective studies. Such studies are required to bring precision medicine to the bedside of critically ill patients with sepsis.
Conflict of interest statement
H.R.W. and the Cincinnati Children’s Hospital Research Foundation hold United States Patents for the PERSEVERE biomarkers and paediatric endotyping strategies described in the manuscript. N.L.S. declares no competing interests.
Figures
References
-
- Marshall JC. Why have clinical trials in sepsis failed? Trends Mol. Med. 2014;20:195–203. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

