Targeting transcription factors in cancer - from undruggable to reality
- PMID: 31511663
- PMCID: PMC8820243
- DOI: 10.1038/s41568-019-0196-7
Targeting transcription factors in cancer - from undruggable to reality
Abstract
Mutated or dysregulated transcription factors represent a unique class of drug targets that mediate aberrant gene expression, including blockade of differentiation and cell death gene expression programmes, hallmark properties of cancers. Transcription factor activity is altered in numerous cancer types via various direct mechanisms including chromosomal translocations, gene amplification or deletion, point mutations and alteration of expression, as well as indirectly through non-coding DNA mutations that affect transcription factor binding. Multiple approaches to target transcription factor activity have been demonstrated, preclinically and, in some cases, clinically, including inhibition of transcription factor-cofactor protein-protein interactions, inhibition of transcription factor-DNA binding and modulation of levels of transcription factor activity by altering levels of ubiquitylation and subsequent proteasome degradation or by inhibition of regulators of transcription factor expression. In addition, several new approaches to targeting transcription factors have recently emerged including modulation of auto-inhibition, proteolysis targeting chimaeras (PROTACs), use of cysteine reactive inhibitors, targeting intrinsically disordered regions of transcription factors and combinations of transcription factor inhibitors with kinase inhibitors to block the development of resistance. These innovations in drug development hold great promise to yield agents with unique properties that are likely to impact future cancer treatment.
Conflict of interest statement
Competing Interests
J.H.B. has a licensing agreement with Systems Oncology for the CBFβ–SMMHC inhibitor AI-10-49 (LeuSO).
Figures

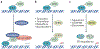

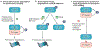
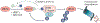
References
-
- Darnell JE Jr. Transcription factors as targets for cancer therapy. Nat. Rev Cancer 2, 740–749 (2002). - PubMed
-
- Arkin MR, Tang Y & Wells JA Small-molecule inhibitors of protein–protein interactions: progressing toward the reality. Chem. Biol 21, 1102–1114 (2014). - PMC - PubMed
-
This comprehensive review illustrates the extraordinary progress that has been made in the development of protein–protein interaction inhibitors for use in a wide variety of disease settings.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

