SLC19A1 transports immunoreactive cyclic dinucleotides
- PMID: 31511694
- PMCID: PMC6785039
- DOI: 10.1038/s41586-019-1553-0
SLC19A1 transports immunoreactive cyclic dinucleotides
Erratum in
-
Author Correction: SLC19A1 transports immunoreactive cyclic dinucleotides.Nature. 2020 Mar;579(7800):E12. doi: 10.1038/s41586-020-2064-8. Nature. 2020. PMID: 32144410
Abstract
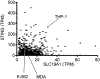
The accumulation of DNA in the cytosol serves as a key immunostimulatory signal associated with infections, cancer and genomic damage1,2. Cytosolic DNA triggers immune responses by activating the cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) pathway3. The binding of DNA to cGAS activates its enzymatic activity, leading to the synthesis of a second messenger, cyclic guanosine monophosphate-adenosine monophosphate (2'3'-cGAMP)4-7. This cyclic dinucleotide (CDN) activates STING8, which in turn activates the transcription factors interferon regulatory factor 3 (IRF3) and nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB), promoting the transcription of genes encoding type I interferons and other cytokines and mediators that stimulate a broader immune response. Exogenous 2'3'-cGAMP produced by malignant cells9 and other CDNs, including those produced by bacteria10-12 and synthetic CDNs used in cancer immunotherapy13,14, must traverse the cell membrane to activate STING in target cells. How these charged CDNs pass through the lipid bilayer is unknown. Here we used a genome-wide CRISPR-interference screen to identify the reduced folate carrier SLC19A1, a folate-organic phosphate antiporter, as the major transporter of CDNs. Depleting SLC19A1 in human cells inhibits CDN uptake and functional responses, and overexpressing SLC19A1 increases both uptake and functional responses. In human cell lines and primary cells ex vivo, CDN uptake is inhibited by folates as well as two medications approved for treatment of inflammatory diseases, sulfasalazine and the antifolate methotrexate. The identification of SLC19A1 as the major transporter of CDNs into cells has implications for the immunotherapeutic treatment of cancer13, host responsiveness to CDN-producing pathogenic microorganisms11 and-potentially-for some inflammatory diseases.
Figures

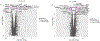
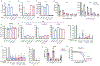










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

