Translational recoding: canonical translation mechanisms reinterpreted
- PMID: 31511883
- PMCID: PMC7026636
- DOI: 10.1093/nar/gkz783
Translational recoding: canonical translation mechanisms reinterpreted
Abstract
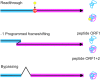
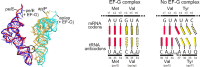
During canonical translation, the ribosome moves along an mRNA from the start to the stop codon in exact steps of one codon at a time. The collinearity of the mRNA and the protein sequence is essential for the quality of the cellular proteome. Spontaneous errors in decoding or translocation are rare and result in a deficient protein. However, dedicated recoding signals in the mRNA can reprogram the ribosome to read the message in alternative ways. This review summarizes the recent advances in understanding the mechanisms of three types of recoding events: stop-codon readthrough, -1 ribosome frameshifting and translational bypassing. Recoding events provide insights into alternative modes of ribosome dynamics that are potentially applicable to other non-canonical modes of prokaryotic and eukaryotic translation.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Joshi K., Cao L., Farabaugh P.. The problem of genetic code misreading during protein synthesis. Yeast. 2019; 36:35–42. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

