Development of highly efficient protocols for extraction and amplification of cytomegalovirus DNA from dried blood spots for detection and genotyping of polymorphic immunomodulatory genes
- PMID: 31513621
- PMCID: PMC6742235
- DOI: 10.1371/journal.pone.0222053
Development of highly efficient protocols for extraction and amplification of cytomegalovirus DNA from dried blood spots for detection and genotyping of polymorphic immunomodulatory genes
Abstract
Congenital cytomegalovirus (CMV) infection is a major cause of birth defects ranging from developmental disorders to stillbirth. Most newborns affected by CMV do not present with symptoms at birth but are at risk of sequelae at later stages of their childhood. Stored dried blood spots (DBS) taken at birth can be used for retrospective diagnosis of hereditary diseases, but detection of pathogens is challenged by potentially low pathogen concentrations in the small blood volume available in a DBS. Here we test four different extraction methods for optimal recovery of CMV DNA from DBS at low to high CMV titers. The recovery efficiencies varied widely between the different extractions (from 3% to 100%) with the most efficient method extracting up to 113-fold more CMV DNA than the least efficient and 8-fold more than the reference protocol. Furthermore, we amplified four immunomodulatory CMV genes from the extracted DNA: the UL40 and UL111A genes which occur as functional knockouts in some circulating CMV strains, and the highly variable UL146 and US28 genes. The PCRs specifically amplified the CMV genes at all tested titers with sufficient quality for sequencing and genotyping. In summary, we here report an extraction method for optimal recovery of CMV DNA from DBSs that can be used for both detection of CMV and for genotyping of polymorphic CMV genes in congenital CMV infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


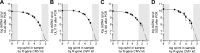

References
-
- Mocarski ES Jr., Immunomodulation by cytomegaloviruses: manipulative strategies beyond evasion. Trends Microbiol. 2002;10(7):332–9. . - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

