The opportunistic pathogen Stenotrophomonas maltophilia utilizes a type IV secretion system for interbacterial killing
- PMID: 31513674
- PMCID: PMC6759196
- DOI: 10.1371/journal.ppat.1007651
The opportunistic pathogen Stenotrophomonas maltophilia utilizes a type IV secretion system for interbacterial killing
Abstract
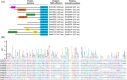
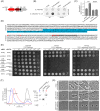
Bacterial type IV secretion systems (T4SS) are a highly diversified but evolutionarily related family of macromolecule transporters that can secrete proteins and DNA into the extracellular medium or into target cells. It was recently shown that a subtype of T4SS harboured by the plant pathogen Xanthomonas citri transfers toxins into target cells. Here, we show that a similar T4SS from the multi-drug-resistant opportunistic pathogen Stenotrophomonas maltophilia is proficient in killing competitor bacterial species. T4SS-dependent duelling between S. maltophilia and X. citri was observed by time-lapse fluorescence microscopy. A bioinformatic search of the S. maltophilia K279a genome for proteins containing a C-terminal domain conserved in X. citri T4SS effectors (XVIPCD) identified twelve putative effectors and their cognate immunity proteins. We selected a putative S. maltophilia effector with unknown function (Smlt3024) for further characterization and confirmed that it is indeed secreted in a T4SS-dependent manner. Expression of Smlt3024 in the periplasm of E. coli or its contact-dependent delivery via T4SS into E. coli by X. citri resulted in reduced growth rates, which could be counteracted by expression of its cognate inhibitor Smlt3025 in the target cell. Furthermore, expression of the VirD4 coupling protein of X. citri can restore the function of S. maltophilia ΔvirD4, demonstrating that effectors from one species can be recognized for transfer by T4SSs from another species. Interestingly, Smlt3024 is homologous to the N-terminal domain of large Ca2+-binding RTX proteins and the crystal structure of Smlt3025 revealed a topology similar to the iron-regulated protein FrpD from Neisseria meningitidis which has been shown to interact with the RTX protein FrpC. This work expands our current knowledge about the function of bacteria-killing T4SSs and increases the panel of effectors known to be involved in T4SS-mediated interbacterial competition, which possibly contribute to the establishment of S. maltophilia in clinical and environmental settings.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Garcia-Bayona L, Comstock LE (2018). Bacterial antagonism in host-associated microbial communities. Science 361. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

