Structural Insights into the Regulation Mechanism of Small GTPases by GEFs
- PMID: 31514408
- PMCID: PMC6767298
- DOI: 10.3390/molecules24183308
Structural Insights into the Regulation Mechanism of Small GTPases by GEFs
Abstract
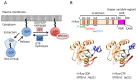
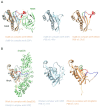
Small GTPases are key regulators of cellular events, and their dysfunction causes many types of cancer. They serve as molecular switches by cycling between inactive guanosine diphosphate (GDP)-bound and active guanosine triphosphate (GTP)-bound states. GTPases are deactivated by GTPase-activating proteins (GAPs) and are activated by guanine-nucleotide exchange factors (GEFs). The intrinsic GTP hydrolysis activity of small GTPases is generally low and is accelerated by GAPs. GEFs promote GDP dissociation from small GTPases to allow for GTP binding, which results in a conformational change of two highly flexible segments, called switch I and switch II, that enables binding of the gamma phosphate and allows small GTPases to interact with downstream effectors. For several decades, crystal structures of many GEFs and GAPs have been reported and have shown tremendous structural diversity. In this review, we focus on the latest structural studies of GEFs. Detailed pictures of the variety of GEF mechanisms at atomic resolution can provide insights into new approaches for drug discovery.
Keywords: GEF; crystal structure; local protein unfolding; regulation mechanism; small GTPases.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures










References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

