The Soybean Laccase Gene Family: Evolution and Possible Roles in Plant Defense and Stem Strength Selection
- PMID: 31514462
- PMCID: PMC6770974
- DOI: 10.3390/genes10090701
The Soybean Laccase Gene Family: Evolution and Possible Roles in Plant Defense and Stem Strength Selection
Abstract
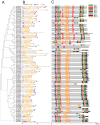
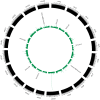
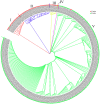
Laccase is a widely used industrial oxidase for food processing, dye synthesis, paper making, and pollution remediation. At present, laccases used by industries come mainly from fungi. Plants contain numerous genes encoding laccase enzymes that show properties which are distinct from that of the fungal laccases. These plant-specific laccases may have better potential for industrial purposes. The aim of this work was to conduct a genome-wide search for the soybean laccase genes and analyze their characteristics and specific functions. A total of 93 putative laccase genes (GmLac) were identified from the soybean genome. All 93 GmLac enzymes contain three typical Cu-oxidase domains, and they were classified into five groups based on phylogenetic analysis. Although adjacent members on the tree showed highly similar exon/intron organization and motif composition, there were differences among the members within a class for both conserved and differentiated functions. Based on the expression patterns, some members of laccase were expressed in specific tissues/organs, while some exhibited a constitutive expression pattern. Analysis of the transcriptome revealed that some laccase genes might be involved in providing resistance to oomycetes. Analysis of the selective pressures acting on the laccase gene family in the process of soybean domestication revealed that 10 genes could have been under artificial selection during the domestication process. Four of these genes may have contributed to the transition of the soft and thin stem of wild soybean species into strong, thick, and erect stems of the cultivated soybean species. Our study provides a foundation for future functional studies of the soybean laccase gene family.
Keywords: Cu-oxidase; GmLac; laccase; resistance; selection; soybean; stem strength.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Evolutionary divergence of function and expression of laccase genes in plants.J Genet. 2020;99:23. J Genet. 2020. PMID: 32366734
-
Genome-wide identification and evolutionary analysis of leucine-rich repeat receptor-like protein kinase genes in soybean.BMC Plant Biol. 2016 Mar 2;16:58. doi: 10.1186/s12870-016-0744-1. BMC Plant Biol. 2016. PMID: 26935840 Free PMC article.
-
Genome-wide survey of the soybean GATA transcription factor gene family and expression analysis under low nitrogen stress.PLoS One. 2015 Apr 17;10(4):e0125174. doi: 10.1371/journal.pone.0125174. eCollection 2015. PLoS One. 2015. PMID: 25886477 Free PMC article.
-
Basic and applied features of multicopper oxidases, CueO, bilirubin oxidase, and laccase.Chem Rec. 2007;7(4):220-9. doi: 10.1002/tcr.20125. Chem Rec. 2007. PMID: 17663447 Review.
-
Laccase Properties, Physiological Functions, and Evolution.Int J Mol Sci. 2020 Jan 31;21(3):966. doi: 10.3390/ijms21030966. Int J Mol Sci. 2020. PMID: 32024019 Free PMC article. Review.
Cited by
-
Comparative transcriptome profiling of resistant and susceptible foxtail millet responses to Sclerospora graminicola infection.BMC Plant Biol. 2022 Dec 6;22(1):567. doi: 10.1186/s12870-022-03963-5. BMC Plant Biol. 2022. PMID: 36471245 Free PMC article.
-
Substrate Specificity of LACCASE8 Facilitates Polymerization of Caffeyl Alcohol for C-Lignin Biosynthesis in the Seed Coat of Cleome hassleriana.Plant Cell. 2020 Dec;32(12):3825-3845. doi: 10.1105/tpc.20.00598. Epub 2020 Oct 9. Plant Cell. 2020. PMID: 33037146 Free PMC article.
-
Plant Copper Metalloenzymes As Prospects for New Metabolism Involving Aromatic Compounds.Front Plant Sci. 2021 Nov 29;12:692108. doi: 10.3389/fpls.2021.692108. eCollection 2021. Front Plant Sci. 2021. PMID: 34925392 Free PMC article. Review.
-
Genome-Wide Identification and Comprehensive Analysis of the PPO Gene Family in Glycine max and Glycine soja.Genes (Basel). 2024 Dec 26;16(1):17. doi: 10.3390/genes16010017. Genes (Basel). 2024. PMID: 39858564 Free PMC article.
-
A Sustainable Approach for Degradation of Alternariol by Peroxidase Extracted from Soybean Hulls: Performance, Pathway, and Toxicity Evaluation.Foods. 2024 Aug 1;13(15):2434. doi: 10.3390/foods13152434. Foods. 2024. PMID: 39123625 Free PMC article.
References
-
- Yaropolov A.I., Skorobogat O.V., Vartanov S.S., Varfolomeyev S.D. Laccase. Appl. Biochem. Biotechnol. 1994;49:257–280. doi: 10.1007/BF02783061. - DOI
-
- Alfred M.M., Richard C.S. Laccase: New functions for an old enzyme. Phytochemistry. 2002;60:551–565. - PubMed
-
- Diamantidis G., Effosse A., Potier P., René B. Purification and characterization of the first bacterial laccase in the rhizospheric bacterium Azospirillum lipoferum. Soil Biol. Biochem. 2000;32:919–927. doi: 10.1016/S0038-0717(99)00221-7. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

