COMMD1 and PtdIns(4,5)P2 interaction maintain ATP7B copper transporter trafficking fidelity in HepG2 cells
- PMID: 31515276
- PMCID: PMC6803360
- DOI: 10.1242/jcs.231753
COMMD1 and PtdIns(4,5)P2 interaction maintain ATP7B copper transporter trafficking fidelity in HepG2 cells
Abstract
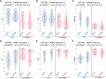
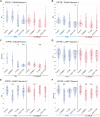
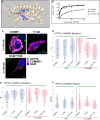
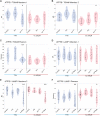
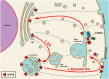
Copper-responsive intracellular ATP7B trafficking is crucial for maintaining the copper balance in mammalian hepatocytes and thus copper levels in organs. The copper metabolism domain-containing protein 1 (COMMD1) binds both the ATP7B copper transporter and phosphatidylinositol (4,5)-bisphosphate [PtdIns(4,5)P2], whereas COMMD1 loss causes hepatocyte copper accumulation. Although it is clear that COMMD1 is localized to endocytic trafficking complexes, a direct function for COMMD1 in ATP7B trafficking has not yet been defined. In this study, experiments using quantitative colocalization analysis reveal that COMMD1 modulates copper-responsive ATP7B trafficking through recruitment to PtdIns(4,5)P2 Decreased COMMD1 abundance results in loss of ATP7B from lysosomes and the trans-Golgi network (TGN) in high copper conditions, although excess expression of COMMD1 also disrupts ATP7B trafficking and TGN structure. Overexpression of COMMD1 mutated to inhibit PtdIns(4,5)P2 binding has little impact on ATP7B trafficking. A mechanistic PtdIns(4,5)P2-mediated function for COMMD1 is proposed that is consistent with decreased cellular copper export as a result of disruption of the ATP7B trafficking itinerary and early endosome accumulation when COMMD1 is depleted. PtdIns(4,5)P2 interaction with COMMD1 as well as COMMD1 abundance could both be important in maintenance of specific membrane protein trafficking pathways.
Keywords: ATP7B; COMMD1; Endosome; Lysosome; Trafficking.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures



References
-
- Bartuzi P., Billadeau D. D., Favier R., Rong S., Dekker D., Fedoseienko A., Fieten H., Wijers M., Levels J. H., Huijkman N. et al. (2016). CCC- and WASH-mediated endosomal sorting of LDLR is required for normal clearance of circulating LDL. Nat. Commun. 7, 10961 10.1038/ncomms10961 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

